4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4CLC
 
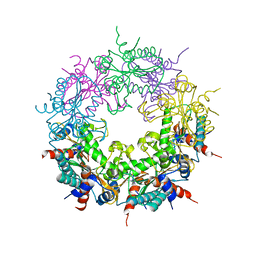 | | Crystal structure of Ybr137w protein | | Descriptor: | UPF0303 PROTEIN YBR137W | | Authors: | Yeh, Y.-H, Lin, T.-W, Lin, C.-Y, Hsiao, C.-D. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Characterization of Ybr137Wp Implicate its Involvement in the Targeting of Tail-Anchored Proteins to Membranes.
Mol.Cell.Biol., 34, 2014
|
|
5GXI
 
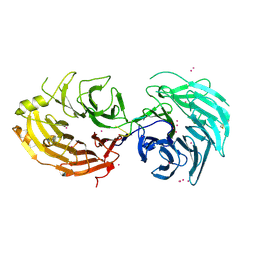 | | Structure of the Gemin5 WD40 domain in complex with AAUUUUUGAG | | Descriptor: | Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*GP*AP*G)-3'), UNKNOWN ATOM OR ION | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
7JYA
 
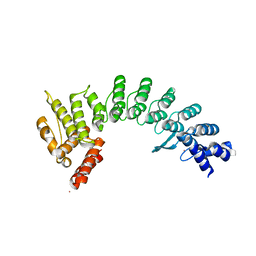 | | Crystal structure of E3 ligase in complex with peptide | | Descriptor: | ASN-ARG-ARG-ARG-ARG-TRP-ARG-GLU-ARG-GLN-ARG, Protein fem-1 homolog C, UNKNOWN ATOM OR ION | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
8WWW
 
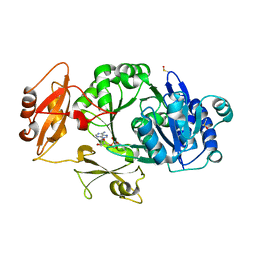 | | Crystal structure of the adenylation domain of SyAstC | | Descriptor: | Carrier domain-containing protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Li, Z, Zhu, Z, Zhu, D, Shen, Y. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Functional Application of the Single-Module NRPS-like d-Alanyltransferase in Maytansinol Biosynthesis
Acs Catalysis, 14, 2024
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
1HS5
 
 | | NMR SOLUTION STRUCTURE OF DESIGNED P53 DIMER | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Davison, T.S, Nie, X, Ma, W, Li, Y, Kay, C, Benchimol, S, Arrowsmith, C.H. | | Deposit date: | 2000-12-22 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functionality of a designed p53 dimer.
J.Mol.Biol., 307, 2001
|
|
1THG
 
 | | 1.8 ANGSTROMS REFINED STRUCTURE OF THE LIPASE FROM GEOTRICHUM CANDIDUM | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrag, J.D, Cygler, M. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A refined structure of the lipase from Geotrichum candidum.
J.Mol.Biol., 230, 1993
|
|
9INS
 
 | |
7PD4
 
 | |
7PD8
 
 | |
7YKS
 
 | |
7PDH
 
 | |
7PDD
 
 | |
7PDG
 
 | |
7PDE
 
 | |
7PDF
 
 | |
7YKR
 
 | |
6EQB
 
 | | HLA class I histocompatibility antigen | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | TCR-induced alteration of primary MHC peptide anchor residue.
Eur.J.Immunol., 49, 2019
|
|
6EAY
 
 | |
6EQA
 
 | | HLA class I histocompatibility antigen | | Descriptor: | ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | TCR-induced alteration of primary MHC peptide anchor residue.
Eur.J.Immunol., 49, 2019
|
|
5I86
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02778174 | | Descriptor: | (4R)-N-benzyl-4-methyl-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepine-6-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
6L4S
 
 | |
5I8B
 
 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
