4Z61
 
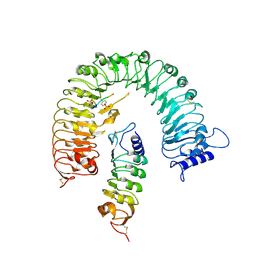 | | The plant peptide hormone receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-ILE-PTR-THR-GLN, Phytosulfokine receptor 1, ... | | Authors: | Chai, J, Wang, J. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
5XKN
 
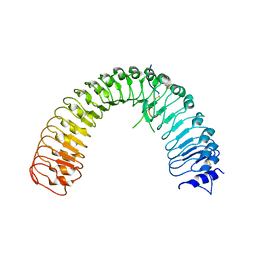 | | Crystal structure of plant receptor ERL2 in complexe with EPFL4 | | Descriptor: | EPIDERMAL PATTERNING FACTOR-like protein 4, LRR receptor-like serine/threonine-protein kinase ERL2 | | Authors: | Chai, J, Lin, G. | | Deposit date: | 2017-05-08 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
5XKJ
 
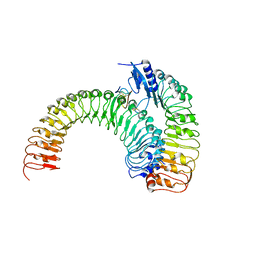 | | Crystal structure of plant receptor ERL1-TMM in complexe with EPF2 | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein EPIDERMAL PATTERNING FACTOR 2, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G. | | Deposit date: | 2017-05-07 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.475 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
4EBZ
 
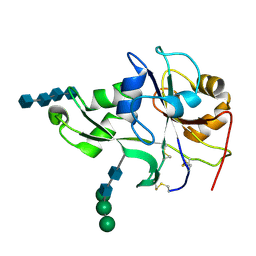 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
4EBY
 
 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | Authors: | Li, H, Wu, Y, Liang, H, Liu, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3MBF
 
 | |
7MLH
 
 | | Crystal structure of human IgE (2F10) in complex with Der p 2.0103 | | Descriptor: | CHLORIDE ION, Der p 2 variant 3, IgE Heavy chain, ... | | Authors: | Khatri, K, Kapingidza, A.B, Richardson, C.M, Vailes, L.D, Wunschmann, S, Dolamore, C, Chapman, M.D, Smith, S.A, Pomes, A, Chruszcz, M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human IgE monoclonal antibody recognition of mite allergen Der p 2 defines structural basis of an epitope for IgE cross-linking and anaphylaxis in vivo.
Pnas Nexus, 1, 2022
|
|
5KZB
 
 | |
2BAX
 
 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | Deposit date: | 2005-10-15 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
8GTP
 
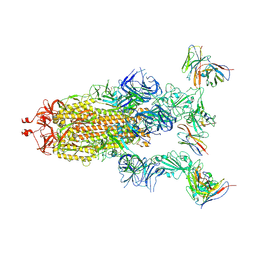 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTO
 
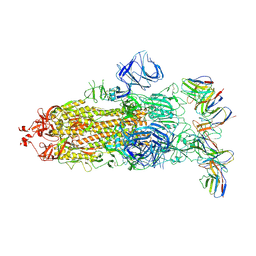 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTQ
 
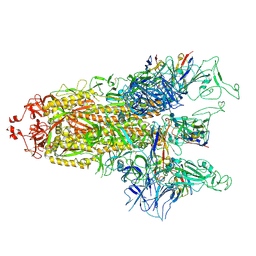 | | cryo-EM structure of Omicron BA.5 S protein in complex with S2L20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
4V9F
 
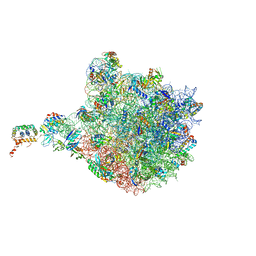 | |
4W6E
 
 | | Human Tankyrase 1 with small molecule inhibitor | | Descriptor: | 2-(4-{6-[(3S)-3,4-dimethylpiperazin-1-yl]-4-methylpyridin-3-yl}phenyl)-8-(hydroxymethyl)quinazolin-4(3H)-one, Tankyrase-1, ZINC ION | | Authors: | Kazmirski, S.L, Johannes, J, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-20 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
5ZLF
 
 | | CRYSTAL STRUCTURE OF OCTAPRENYL PYROPHOSPHATE SYNTHASE FROM ESCHERICHIA COLI WITH ligand BPH-629 | | Descriptor: | MAGNESIUM ION, Octaprenyl diphosphate synthase, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-27 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
5ZE6
 
 | | CRYSTAL STRUCTURE OF OCTAPRENYL PYROPHOSPHATE SYNTHASE FROM ESCHERICHIA COLI WITH BPH-981 | | Descriptor: | 2-hydroxy-6-(tetradecyloxy)benzoic acid, MAGNESIUM ION, Octaprenyl diphosphate synthase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-02-26 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
8HAW
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8HAV
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8H3X
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8H3Y
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 327 | | Descriptor: | Fragilysin, Nanobody 327, ZINC ION | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
5ZHE
 
 | | STRUCTURE OF E. COLI UNDECAPRENYL DIPHOSPHATE SYNTHASE IN COMPLEX WITH BPH-981 | | Descriptor: | 2-hydroxy-6-(tetradecyloxy)benzoic acid, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
5UQ9
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase with ((4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl)methyl dihydrogen phosphate | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating, [(4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl]methyl dihydrogen phosphate | | Authors: | Leonard, P.G. | | Deposit date: | 2017-02-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation.
Cell Rep, 26, 2019
|
|
8SJ7
 
 | |
