6Q9J
 
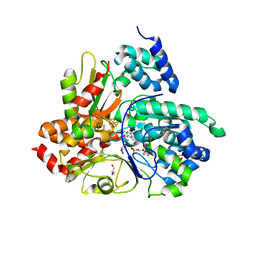 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129S and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6C1Y
 
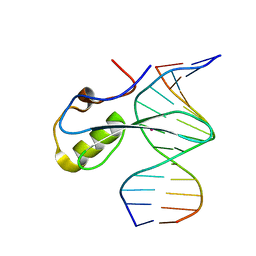 | | mbd of human mecp2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Bian, C, Tempel, W, Wernimont, A.K, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
6XZ5
 
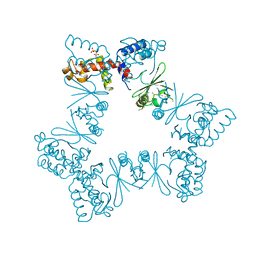 | |
2AV8
 
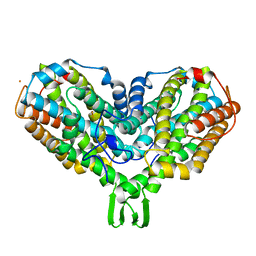 | | Y122F MUTANT OF RIBONUCLEOTIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | FE (II) ION, MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
6Q9K
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF S96M bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
7SR6
 
 | | Human Endogenous Retrovirus (HERV-K) reverse transcriptase ternary complex with dsDNA template Primer and dNTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Baldwin, E.T, Nichols, C. | | Deposit date: | 2021-11-08 | | Release date: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K-specific inhibition.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5GMJ
 
 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-B C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule B | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.986 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
6JQH
 
 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
4IO0
 
 | | Crystal structure of F128A mutant of an epoxide hydrolase from Bacillus megaterium complexed with its product (R)-3-[1]naphthyloxy-propane-1,2-diol | | Descriptor: | (2R)-3-(naphthalen-1-yloxy)propane-1,2-diol, SULFATE ION, Soluble epoxide hydrolase | | Authors: | Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2013-01-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4YZ9
 
 | |
4YZC
 
 | | Crystal structure of pIRE1alpha in complex with staurosporine | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMR
 
 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
1ZX8
 
 | |
6IIV
 
 | | Crystal structure of the human thromboxane A2 receptor bound to daltroban | | Descriptor: | 2-[4-[2-[(4-chlorophenyl)sulfonylamino]ethyl]phenyl]ethanoic acid, CHOLESTEROL, GLYCEROL, ... | | Authors: | Fan, H, Zhao, Q, Wu, B. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for ligand recognition of the human thromboxane A2receptor.
Nat. Chem. Biol., 15, 2019
|
|
5GMI
 
 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-C C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule C | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
7X4I
 
 | |
1ZKG
 
 | |
2B1B
 
 | |
6MBZ
 
 | | Structure of Transcription Factor | | Descriptor: | Signal transducer and activator of transcription 5B | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-08-30 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural and functional consequences of the STAT5BN642Hdriver mutation.
Nat Commun, 10, 2019
|
|
8SUF
 
 | | The complex of TOL-1 ectodomain bound to LAT-1 Lectin domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Latrophilin-like protein 1, ... | | Authors: | Carmona Rosas, G, Li, J, Arac, D, Ozkan, E. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis and functional roles for Toll-like receptor binding to Latrophilin adhesion-GPCR in embryo development
To Be Published
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
6MBW
 
 | | Structure of Transcription Factor | | Descriptor: | Signal transducer and activator of transcription 5B | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-08-30 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural and functional consequences of the STAT5BN642H driver mutation.
Nat Commun, 10, 2019
|
|
