4QO8
 
 | | Lactate Dehydrogenase A in complex with substituted 3-Hydroxy-2-mercaptocyclohex-2-enone compound 104 | | Descriptor: | (5S)-2-[(2-chlorophenyl)sulfanyl]-5-(2,6-dichlorophenyl)-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Identification of substituted 3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4QO7
 
 | |
5YHQ
 
 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
4R69
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 13 | | Descriptor: | (5R)-2-[(2-chlorophenyl)sulfanyl]-5-[2,6-dichloro-3-(tetrahydro-2H-pyran-4-ylamino)phenyl]-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
4RE1
 
 | | Crystal structure of human TEAD1 and disulfide-engineered YAP | | Descriptor: | CHLORIDE ION, Transcriptional enhancer factor TEF-1, Yorkie homolog | | Authors: | Xu, Z, Zhou, Z. | | Deposit date: | 2014-09-21 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting Hippo pathway by specific interruption of YAP-TEAD interaction using cyclic YAP-like peptides.
Faseb J., 29, 2015
|
|
6A5F
 
 | |
4PSX
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
1D6E
 
 | | CRYSTAL STRUCTURE OF HLA-DR4 COMPLEX WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, PEPTIDOMIMETIC INHIBITOR | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-13 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
6A5H
 
 | |
4R68
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 31 | | Descriptor: | (1S)-1-phenylethyl (4-chloro-3-{[(4S)-4-(2,6-dichlorophenyl)-2-hydroxy-6-oxocyclohex-1-en-1-yl]sulfanyl}phenyl)acetate, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
1D5M
 
 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDE AND SEB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Swain, A.L, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-07 | | Release date: | 2000-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
6A5G
 
 | |
1D5X
 
 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH DIPEPTIDE MIMETIC AND SEB | | Descriptor: | DIPEPTIDE MIMETIC INHIBITOR, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
5U6I
 
 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
4KDC
 
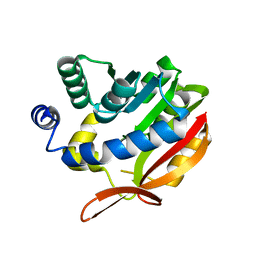 | | Crystal Structure of UBIG | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins.
Biochem. J., 470, 2015
|
|
8I8A
 
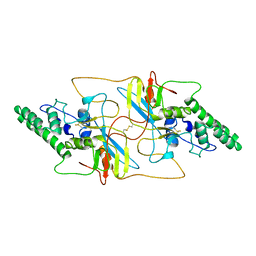 | |
8I8C
 
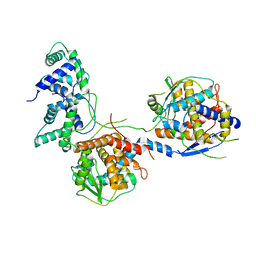 | |
8I8B
 
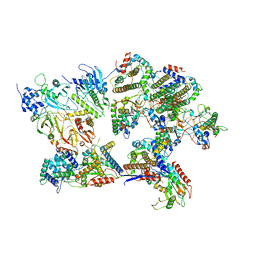 | | Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) | | Descriptor: | 38K, AcOrf-109 peptide, Early 49 Daa protein, ... | | Authors: | Jia, X, Gao, Y, Zhang, Q. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Architecture of the baculovirus nucleocapsid revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
6CWA
 
 | |
4Q77
 
 | | Crystal structure of Rot, a global regulator of virulence genes in Staphylococcus aureus | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator rot | | Authors: | Zhu, Y, Fan, X, Li, X, Teng, M. | | Deposit date: | 2014-04-24 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of Rot, a global regulator of virulence genes in Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YAD
 
 | | Crystal structure of Marf1 Lotus domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1, SULFATE ION | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YAA
 
 | | Crystal structure of Marf1 NYN domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1 | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1O3S
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: Alteration of DNA Binding Specificity Through Alteration of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
5ZJI
 
 | | Structure of photosystem I supercomplex with light-harvesting complexes I and II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Ma, J, Su, X.D, Cao, P, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II.
Science, 360, 2018
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
