8EX5
 
 | |
8EX6
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1*) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX4
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX7
 
 | |
8EX8
 
 | |
5DQU
 
 | | Crystal Structure of Cas-DNA-10 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
5DQT
 
 | | Crystal Structure of Cas-DNA-22 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (33-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
5DQZ
 
 | | Crystal Structure of Cas-DNA-PAM complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (36-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
5DLJ
 
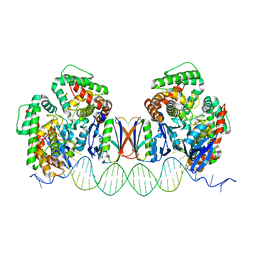 | | Crystal Structure of Cas-DNA-N1 complex | | Descriptor: | 39-mer DNA N1-F, 39-mer DNA N1-R, CRISPR-associated endonuclease Cas1, ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-05 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
2P2H
 
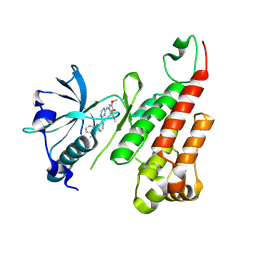 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridinyl-triazine inhibitor | | Descriptor: | 4-(2-anilinopyridin-3-yl)-N-(3,4,5-trimethoxyphenyl)-1,3,5-triazin-2-amine, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of a Highly Selective and Potent 2-(Pyridin-2-yl)-1,3,5-triazine Tie-2 Kinase Inhibitor
J.Med.Chem., 50, 2007
|
|
9G64
 
 | | F420-dependent oxidoreductase | | Descriptor: | F420-dependent oxidoreductase | | Authors: | Mueller, R, Zhao, H, Sikandar, A. | | Deposit date: | 2024-07-17 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Tandem ketone reduction in pepstatin biosynthesis reveals an F 420 H 2 -dependent statine pathway.
Nat Commun, 16, 2025
|
|
9GKH
 
 | |
9ILS
 
 | |
9ILR
 
 | | The structure of the GmvT-GmvA-AcCoA ternary complex | | Descriptor: | ACETYL COENZYME *A, DUF1778 domain-containing protein, N-acetyltransferase, ... | | Authors: | Xie, W, Chen, R, Zhao, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the Shigella flexneri GmvAT toxin-antitoxin system.
Febs Lett., 599, 2025
|
|
9GNC
 
 | |
9GND
 
 | |
9GM0
 
 | |
2P2I
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a nicotinamide inhibitor | | Descriptor: | N-(4-phenoxyphenyl)-2-[(pyridin-4-ylmethyl)amino]nicotinamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a Highly Selective and Potent 2-(Pyridin-2-yl)-1,3,5-triazine Tie-2 Kinase Inhibitor
J.Med.Chem., 50, 2007
|
|
2QU6
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzoxazole inhibitor | | Descriptor: | 4-({2-[(4-chloro-3-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}phenyl)amino]-1,3-benzoxazol-5-yl}oxy)-N-methylpyridine-2-carboxamide, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Evaluation of Orally Active Benzimidazoles and Benzoxazoles as Vascular Endothelial Growth Factor-2 Receptor Tyrosine Kinase Inhibitors.
J.Med.Chem., 50, 2007
|
|
9JVP
 
 | | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
9JVZ
 
 | | CryoEM structure of M. tuberculosis ClpP1P2 bound to bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, W, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
2QU5
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzimidazole inhibitor | | Descriptor: | 4-[[2-[[4-chloro-3-(trifluoromethyl)phenyl]amino]-3H-benzimidazol-5-yl]oxy]-N-methyl-pyridine-2-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design, Synthesis, and Evaluation of Orally Active Benzimidazoles and Benzoxazoles as Vascular Endothelial Growth Factor-2 Receptor Tyrosine Kinase Inhibitors.
J.Med.Chem., 50, 2007
|
|
9IV9
 
 | | Cryo-EM structure of a truncated Nipah Virus L Protein bound by Phosphoprotein Tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-07-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
9IVA
 
 | | Cryo-EM structure of the full-length Nipah Virus L Protein bound by Phosphoprotein Tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-07-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
4HNF
 
 | | Crystal structure of ck1d in complex with pf4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform delta | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
