8GWN
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWB
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWE
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, MAGNESIUM ION, Non-structural protein 8, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWF
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.Y, Huang, Y.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GW1
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L, Rao, Z, Lou, Z. | | Deposit date: | 2022-09-16 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWM
 
 | | SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP | | Descriptor: | 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate), Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWK
 
 | | SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
6J8L
 
 | |
7BRO
 
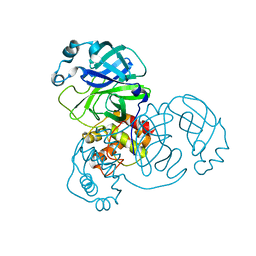 | | Crystal structure of the 2019-nCoV main protease | | Descriptor: | 3C-like proteinase | | Authors: | Fu, L.F. | | Deposit date: | 2020-03-29 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
5CMZ
 
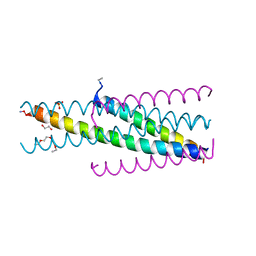 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CN0
 
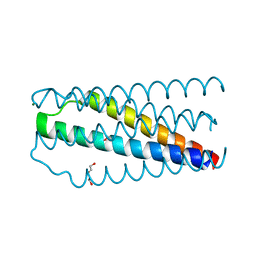 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
7C6S
 
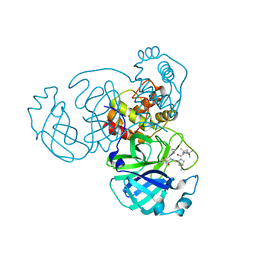 | |
7BRP
 
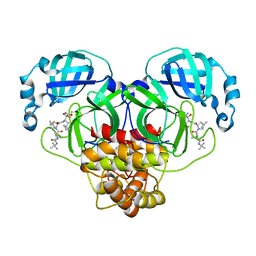 | |
7C6U
 
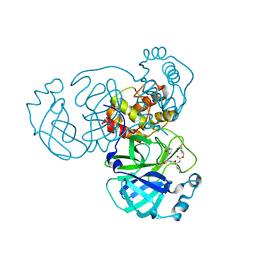 | | Crystal structure of SARS-CoV-2 complexed with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Fu, L, Feng, Y, Qi, J, Gao, F.G. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
6KI2
 
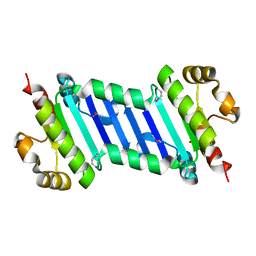 | |
6KI1
 
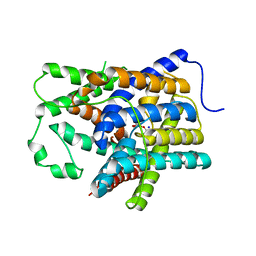 | |
3T0H
 
 | |
3T0Z
 
 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
5H0N
 
 | |
5GUF
 
 | |
8J2M
 
 | | The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constitutively active state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Na+/H+ antiporter | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
8IWO
 
 | | The rice Na+/H+ antiporter SOS1 in an auto-inhibited state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, OsSOS1 | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
3T10
 
 | | HSP90 N-terminal domain bound to ACP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
