6NHP
 
 | |
5VTW
 
 | |
7XGO
 
 | | Human renin in complex with compound2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7XGK
 
 | | Human renin in complex with compound1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7XGP
 
 | | Human renin in complex with compound3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of SPH3127: A Novel, Highly Potent, and Orally Active Direct Renin Inhibitor.
J.Med.Chem., 65, 2022
|
|
7WT9
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab 9A8 | | Descriptor: | Heavy chain of Fab 9A8, Light chain of Fab 9A8, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT7
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT8
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7XPN
 
 | |
7YG7
 
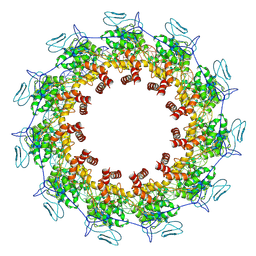 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
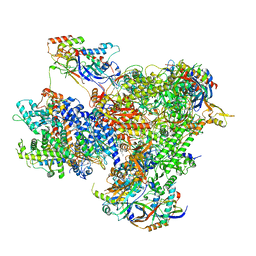 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
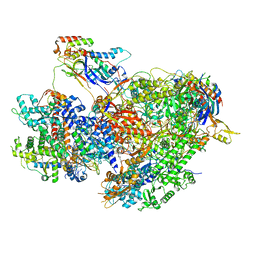 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
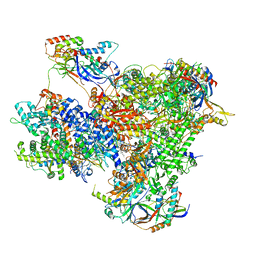 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
4GPI
 
 | | Crystal structure of human B type phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Zhou, L, He, C. | | Deposit date: | 2012-08-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0817 Å) | | Cite: | Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation.
Nat Commun, 4, 2013
|
|
4GPZ
 
 | |
6Y1L
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47R mutant HEAVY CHAIN, FAB A.17 L47R mutant Light CHAIN, ... | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y49
 
 | | Crystal structure of the paraoxon-modified A.17kappa antibody FAB fragment | | Descriptor: | A.17kappa antibody FAB fragment - Heavy Chain, A.17kappa antibody FAB fragment - Light Chain, DIETHYL PHOSPHONATE | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y1N
 
 | | Crystal structure of the phosphonate-modified A.5 antibody FAB fragment | | Descriptor: | 8-METHYL-8-AZABICYCLO[3.2.1]OCTAN-3-YL PHENYLPHOSPHONATE, FAB A.5 Heavy chain, FAB A.5 Light Chain | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y1K
 
 | | Crystal structure of the unmodified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | FAB A.17 L47R mutant Heavy Chain, FAB A.17 L47R mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y1M
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47K mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47K mutant HEAVY CHAIN, FAB A.17 L47K mutant Light CHAIN | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4GWG
 
 | |
4GWK
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase complexed with 3-phosphoglyceric acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PHOSPHOGLYCERIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | He, C, Zhou, L, Zhang, L. | | Deposit date: | 2012-09-03 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth.
Cancer Cell, 22, 2012
|
|
