5C9F
 
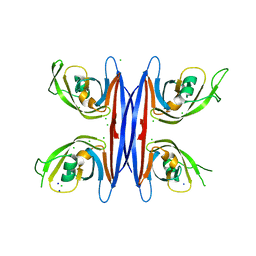 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, CHLORIDE ION, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5E0S
 
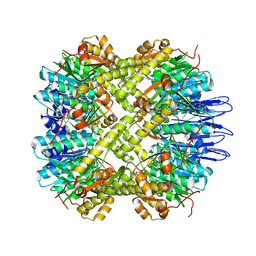 | | crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5DZK
 
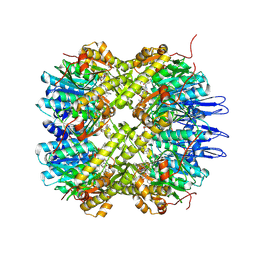 | | Crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, BEZ-LEU-LEU | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5DUY
 
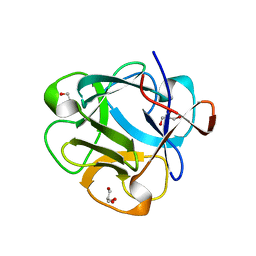 | | Structure of lectin from the sea mussel Crenomytilus grayanus | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Lubkowski, J, Jakob, M, O'Keefe, B, Wlodawer, A. | | Deposit date: | 2015-09-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a lectin from the sea mussel Crenomytilus grayanus (CGL).
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4ZOT
 
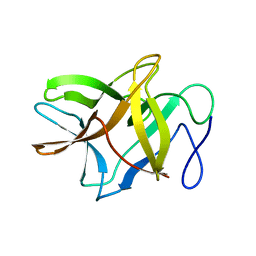 | | Crystal structure of BbKI, a disulfide-free plasma kallikrein inhibitor at 1.4 A resolution | | Descriptor: | Kunitz-type serine protease inhibitor BbKI | | Authors: | Shabalin, I.G, Zhou, D, Wlodawer, A, Oliva, M.L.V. | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of BbKI, a disulfide-free plasma kallikrein inhibitor.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2FIV
 
 | | Crystal structure of feline immunodeficiency virus protease complexed with a substrate | | Descriptor: | ACE-ALN-VAL-STA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-21 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
8R2C
 
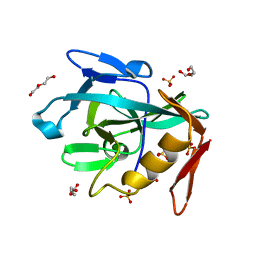 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 598, 2024
|
|
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEN
 
 | | Room temperature X-ray diffraction studies of cisplatin binding to HEWL in DMSO media after 14 months of crystal storage - new refinement | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEO
 
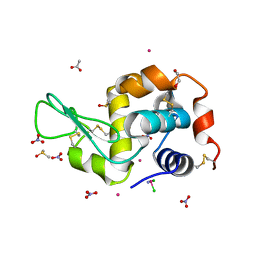 | | Triclinic HEWL co-crystallised with cisplatin, studied at a data collection temperature of 150K - new refinement | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cisplatin, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3IL8
 
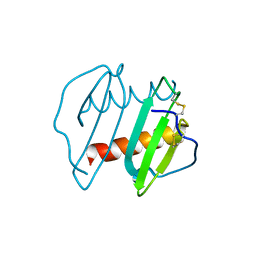 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
4FIV
 
 | | FIV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, FELINE IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-15 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
1B11
 
 | | STRUCTURE OF FELINE IMMUNODEFICIENCY VIRUS PROTEASE COMPLEXED WITH TL-3-093 | | Descriptor: | PROTEIN (Feline Immunodeficiency Virus PROTEASE), SULFATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Gustchina, A, Li, M, Wlodawer, A. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
3C9X
 
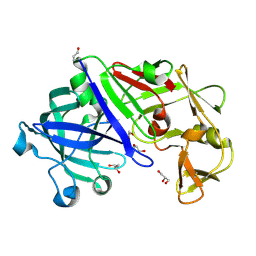 | | Crystal structure of Trichoderma reesei aspartic proteinase | | Descriptor: | GLYCEROL, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-02-19 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
3EMY
 
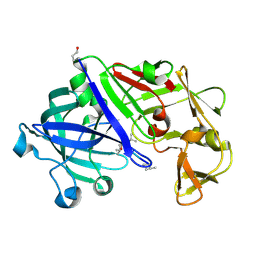 | | Crystal structure of Trichoderma reesei aspartic proteinase complexed with pepstatin A | | Descriptor: | Pepstatin, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal
structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
2NR6
 
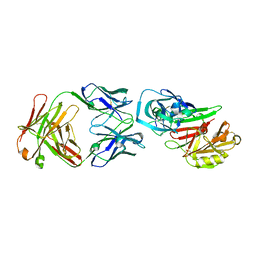 | | Crystal structure of the complex of antibody and the allergen Bla g 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody heavy chain, ... | | Authors: | Li, M, Gustchina, A, Wlodawer, A, Pomes, A, Wunschmann, S. | | Deposit date: | 2006-11-01 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody.
J.Biol.Chem., 283, 2008
|
|
1L6S
 
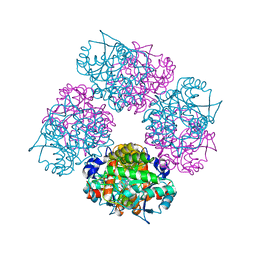 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4,7-Dioxosebacic Acid | | Descriptor: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | Authors: | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-03-13 | | Release date: | 2002-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
1L6Y
 
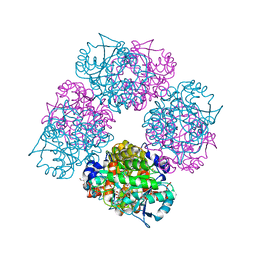 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4-Oxosebacic Acid | | Descriptor: | 4-OXODECANEDIOIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
3D9A
 
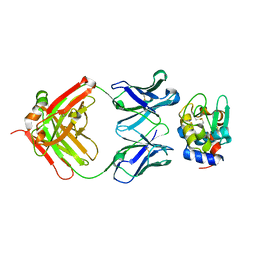 | | High Resolution Crystal Structure Structure of HyHel10 Fab Complexed to Hen Egg Lysozyme | | Descriptor: | Heavy Chain of HyHel10 Antibody Fragment (Fab), Light Chain of HyHel10 Antibody Fragment (Fab), Lysozyme C | | Authors: | DeSantis, M.E, Li, M, Shanmuganathan, A, Acchione, M, Walter, R, Wlodawer, A, Smith-Gill, S. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Light chain somatic mutations change thermodynamics of binding and water coordination in the HyHEL-10 family of antibodies.
Mol.Immunol., 47, 2009
|
|
1ZVJ
 
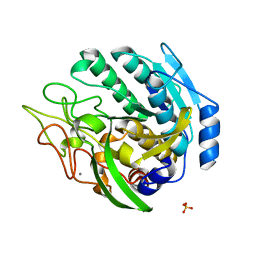 | | Structure of Kumamolisin-AS mutant, D164N | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
2ZNX
 
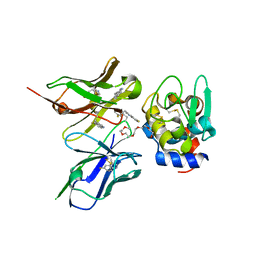 | | 5-Fluorotryptophan Incorporated ScFv10 Complexed to Hen Egg Lysozyme | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lysozyme C, ScFv | | Authors: | DeSantis, M.E, Acchione, M, Li, M, Walter, R.L, Wlodawer, A, Smith-Gill, S. | | Deposit date: | 2008-05-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specific fluorine labeling of the HyHEL10 antibody affects antigen binding and dynamics
Biochemistry, 51, 2012
|
|
2ZNW
 
 | | Crystal Structure of ScFv10 in Complex with Hen Egg Lysozyme | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lysozyme C, ScFv10 | | Authors: | DeSantis, M.E, Acchione, M, Li, M, Walter, R.L, Wlodawer, A, Smith-Gill, S. | | Deposit date: | 2008-05-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Specific fluorine labeling of the HyHEL10 antibody affects antigen binding and dynamics
Biochemistry, 51, 2012
|
|
3FNU
 
 | | Crystal structure of KNI-10006 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, HAP protein | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
8TN7
 
 | | The Crystal Structure of a human monoclonal antibody (aAb), termed TG10, complexed with a disaccharide | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Temme, S, Gildersleeve, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Insights into biofilm architecture and maturation enable improved clinical strategies for exopolysaccharide-targeting therapeutics.
Cell Chem Biol, 31, 2024
|
|
8TN5
 
 | | The Crystal Structure of a human monoclonal antibody (aAb), termed TG10, complexed with a GlcNH2 | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Temme, S, Gildersleeve, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Insights into biofilm architecture and maturation enable improved clinical strategies for exopolysaccharide-targeting therapeutics.
Cell Chem Biol, 31, 2024
|
|
