8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14182 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7YI3
 
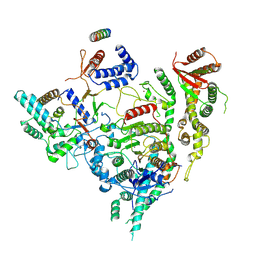 | | Cryo-EM structure of Rpd3S in close-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7YI2
 
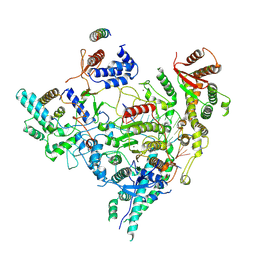 | | Cryo-EM structure of Rpd3S in loose-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI1
 
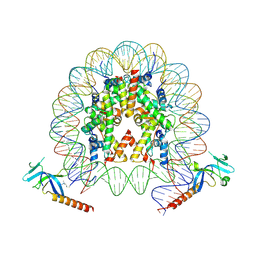 | | Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI0
 
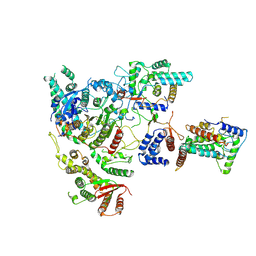 | | Cryo-EM structure of Rpd3S complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI5
 
 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI4
 
 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
6DU5
 
 | |
6DU4
 
 | | Crystal structure of hMettl16 catalytic domain in complex with MAT2A 3'UTR hairpin 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase, ... | | Authors: | Doxtader, K, Wang, P, Nam, Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor.
Mol. Cell, 71, 2018
|
|
1P9U
 
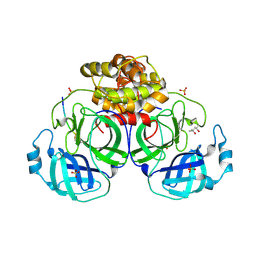 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHQ-VNSTLQ-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION, ... | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
1P9S
 
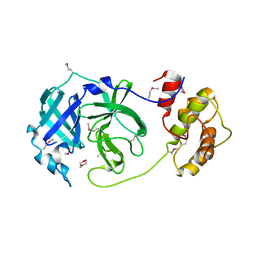 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replicase polyprotein 1ab | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
3SHB
 
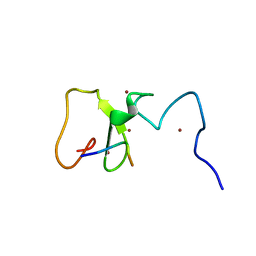 | | Crystal Structure of PHD Domain of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 peptide, ZINC ION | | Authors: | Hu, L, Li, Z, Wang, P, Lin, Y, Xu, Y. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2.
Cell Res., 2011
|
|
3U0J
 
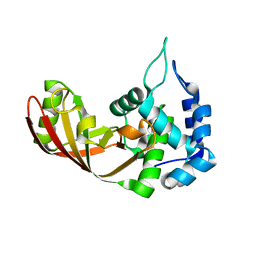 | |
1WNH
 
 | | Crystal structure of mouse Latexin (tissue carboxypeptidase inhibitor) | | Descriptor: | Latexin | | Authors: | Aagaard, A, Listwan, P, Cowieson, N, Huber, T, Ravasi, T, Wells, C.A, Flanagan, J.U, Hume, D.A, Kobe, B, Martin, J.L. | | Deposit date: | 2004-08-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Inflammatory Role for the Mammalian Carboxypeptidase Inhibitor Latexin: Relationship to Cystatins and the Tumor Suppressor TIG1
Structure, 13, 2005
|
|
4OD0
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
6IXL
 
 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase, SULFATE ION | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-11 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
6IXN
 
 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and citrate | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-11 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
5GKQ
 
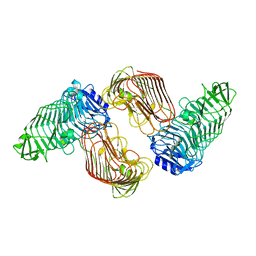 | | Structure of PL6 family alginate lyase AlyGC mutant-R241A | | Descriptor: | AlyGC mutant - R241A, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Zhang, Y.Z, Wang, P, Xu, F. | | Deposit date: | 2016-07-05 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6
J. Biol. Chem., 292, 2017
|
|
5GXD
 
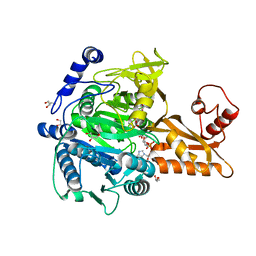 | |
5GKD
 
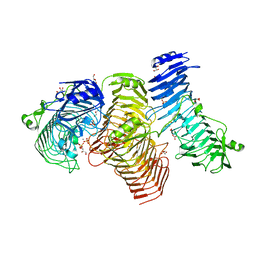 | | Structure of PL6 family alginate lyase AlyGC | | Descriptor: | AlyGC, CALCIUM ION, CARBONATE ION, ... | | Authors: | Zhang, Y.Z, Wang, P, Xu, F. | | Deposit date: | 2016-07-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6
J. Biol. Chem., 292, 2017
|
|
4NM6
 
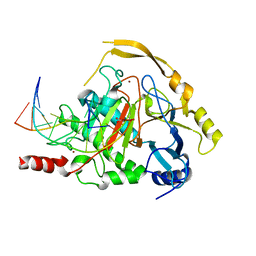 | | Crystal structure of TET2-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*T)-3', FE (II) ION, Methylcytosine dioxygenase TET2, ... | | Authors: | Hu, L, Li, Z, Cheng, J, Rao, Q, Gong, W, Liu, M, Wang, P, Xu, Y. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation.
Cell(Cambridge,Mass.), 155, 2013
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
6IXT
 
 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and Mg2+ | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-12 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
