6PXH
 
 | | Crystal Structure of MERS-CoV S1-NTD bound with G2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIHYDROFOLIC ACID, ... | | Authors: | Wang, N, McLellan, J.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
6PZ8
 
 | | MERS S0 trimer in complex with variable domain of antibody G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G2 heavy chain, ... | | Authors: | Bowman, C.A, Pallesen, J, Ward, A.B. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
6S8Z
 
 | |
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
3NUP
 
 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(1-methylpiperidin-4-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
3NUX
 
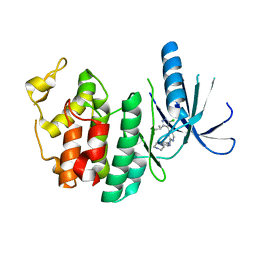 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[5-chloro-3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(5-piperazin-1-ylpyridin-2-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
7TBF
 
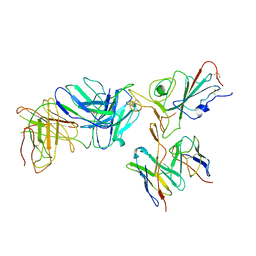 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 antibody A19-61.1, Heavy chain of SARS-CoV-2 antibody B1-182.1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TB8
 
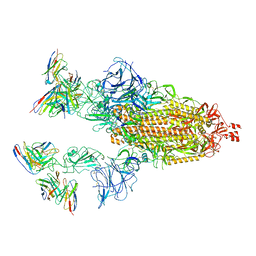 | | Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TCA
 
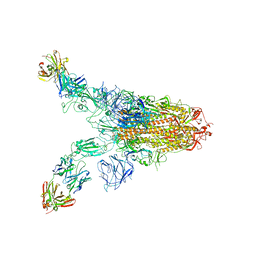 | | Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody A19-46.1, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TCC
 
 | |
7TC9
 
 | |
7TCQ
 
 | |
7S4U
 
 | | Cryo-EM structure of Cas9 in complex with 12-14MM DNA substrate, 5 minute time-point | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand, Target strand, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4V
 
 | | Cas9 bound to 12-14MM DNA, 60 min time-point, kinked conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4X
 
 | | Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
6G1T
 
 | | TraN, a repressor of an Enterococcus conjugative type IV secretion system | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AM32, DNA (34-MER) | | Authors: | Goessweiner-Mohr, N, Kohler, V, Keller, W. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | TraN: A novel repressor of an Enterococcus conjugative type IV secretion system.
Nucleic Acids Res., 46, 2018
|
|
6W52
 
 | | Prefusion RSV F bound by neutralizing antibody RSB1 | | Descriptor: | Fusion glycoprotein F0, Fusion glycoprotein F1 fused with Fibritin trimerization domain, RSB1 Fab Heavy Chain, ... | | Authors: | Harshbarger, W, Chandramouli, S, Malito, M. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Convergent structural features of respiratory syncytial virus neutralizing antibodies and plasticity of the site V epitope on prefusion F.
Plos Pathog., 16, 2020
|
|
6W5D
 
 | | Crystal Structure of Fab RSB1 | | Descriptor: | RSB1 Fab Heavy Chain, RSB1 Fab Light Chain | | Authors: | Harshbarger, W, Chandramouli, S, Malito, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Convergent structural features of respiratory syncytial virus neutralizing antibodies and plasticity of the site V epitope on prefusion F.
Plos Pathog., 16, 2020
|
|
7U0D
 
 | |
4ZPV
 
 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
