7WOQ
 
 | | The state 1 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
5CB8
 
 | |
5CB6
 
 | | Structure of adenosine-5'-phosphosulfate kinase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Herrmann, J, Jez, J.M. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Recapitulating the Structural Evolution of Redox Regulation in Adenosine 5'-Phosphosulfate Kinase from Cyanobacteria to Plants.
J.Biol.Chem., 290, 2015
|
|
4I5N
 
 | | Structural mechanism of trimeric PP2A holoenzyme involving PR70: insight for Cdc6 dephosphorylation | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Microcystin-LR (MCLR) bound form, ... | | Authors: | Wlodarchak, N, Satyshur, K.A, Guo, F, Xing, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
4I5J
 
 | | PP2A PR70 Holoenzyme | | Descriptor: | CALCIUM ION, Serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit alpha | | Authors: | Xing, Y, Jeffery, P.D, Shi, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
5WC8
 
 | |
5WC6
 
 | |
5WD7
 
 | | Structure of a bacterial polysialyltransferase in complex with fondaparinux | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, SULFATE ION, SiaD | | Authors: | Worrall, L.J, Lizak, C, Strynadka, N.C.J. | | Deposit date: | 2017-07-04 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid.
Sci Rep, 7, 2017
|
|
5WCN
 
 | |
4I5K
 
 | |
4I5L
 
 | | Structural mechanism of trimeric PP2A holoenzyme involving PR70: insight for Cdc6 dephosphorylation | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Wlodarchak, N, Satyshur, K.A, Guo, F, Xing, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
5VXA
 
 | | Structure of the human Mesh1-NADPH complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-05-23 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis.
Nat Metab, 2, 2020
|
|
1XX2
 
 | | Refinement of P99 beta-lactamase from Enterobacter cloacae | | Descriptor: | Beta-lactamase | | Authors: | Knox, J.R, Sun, T. | | Deposit date: | 2004-11-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a beta-lactamase transition-state analog.
Biochemistry, 33, 1994
|
|
3AJL
 
 | | Crystal structure of d(CGCGGATf5UCGCG): 5-Formyluridine:guanosine Base-pair in B-DNA with DAPI | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*TP*(UFR)P*CP*GP*CP*G)-3', 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, MAGNESIUM ION | | Authors: | Tsunoda, M, Sakaue, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the structures of DNA damaged by hydroxyl radical: crystal structures of DNA duplexes containing 5-formyluracil
J Nucleic Acids, 2010, 2010
|
|
3AJK
 
 | | Crystal structure of d(CGCGGATf5UCGCG): 5-Formyluridine:Guanosine Base-pair in B-DNA with Hoechst33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*GP*AP*TP*(UFR)P*CP*GP*CP*G*)-3', MAGNESIUM ION | | Authors: | Tsunoda, M, Sakaue, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the structures of DNA damaged by hydroxyl radical: crystal structures of DNA duplexes containing 5-formyluracil
J Nucleic Acids, 2010, 2010
|
|
3AJJ
 
 | | Crystal Structure of d(CGCGGATf5UCGCG): 5-Formyluridine/Guanosine Base-pair in B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*TP*(UFR)P*CP*GP*CP*G)-3' | | Authors: | Tsunoda, M, Sakaue, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Insights into the structures of DNA damaged by hydroxyl radical: crystal structures of DNA duplexes containing 5-formyluracil
J Nucleic Acids, 2010, 2010
|
|
3O3R
 
 | | Crystal Structure of AKR1B14 in complex with NADP | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, Dhagat, U, El-Kabbani, O. | | Deposit date: | 2010-07-26 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of rat aldose reductase-like protein AKR1B14 holoenzyme: Probing the role of His269 in coenzyme binding by site-directed mutagenesis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1KEG
 
 | | Antibody 64M-2 Fab complexed with dTT(6-4)TT | | Descriptor: | 5'-D(*TP*(64T)P*TP*T)-3', Anti-(6-4) photoproduct antibody 64M-2 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-2 Fab (light chain), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Sato, K, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2001-11-15 | | Release date: | 2002-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DNA (6-4) photoproduct dTT(6-4)TT in complex with the 64M-2 antibody Fab fragment implies increased antibody-binding affinity by the flanking nucleotides.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1EHL
 
 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | Descriptor: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
1SD0
 
 | | Structure of arginine kinase C271A mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Gattis, J.L, Ruben, E, Fenley, M.O, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2004-02-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The active site cysteine of arginine kinase: structural and functional analysis of partially active mutants
Biochemistry, 43, 2004
|
|
6LFE
 
 | | Rat-COMT, Nitecapone,SAM and Mg bond | | Descriptor: | 3-(3,4-dihydroxy-5-nitrobenzylidene)pentane-2,4-dione, Catechol O-methyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Catechol O-Methyltransferase Complexed with Nitecapone.
Chem Pharm Bull (Tokyo), 68, 2020
|
|
3QKZ
 
 | | Crystal structure of mutant His269Arg AKR1B14 | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, El-Kabbani, O. | | Deposit date: | 2011-02-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of the His269Arg mutant of the rat aldose reductase-like protein AKR1B14 complexed with NADPH.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1999-04-28 | | Release date: | 1999-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
8BSE
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD) in complex with 1D1 Fab | | Descriptor: | 1D1 FAB HEAVY CHAIN, 1D1 FAB LIGHT CHAIN, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8BSF
 
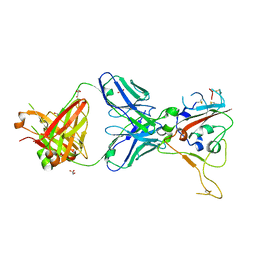 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD-beta variant) in complex with 3D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3D2 FAB HEAVY CHAIN, 3D2 FAB LIGHT CHAIN, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
