5JSM
 
 | |
1GSS
 
 | | THREE-DIMENSIONAL STRUCTURE OF CLASS PI GLUTATHIONE S-TRANSFERASE FROM HUMAN PLACENTA IN COMPLEX WITH S-HEXYLGLUTATHIONE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-hexyl-L-cysteinylglycine | | Authors: | Reinemer, P, Dirr, H.W, Ladenstein, R, Lobello, M, Federici, G, Huber, R, Parker, M.W. | | Deposit date: | 1992-05-28 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
5JT2
 
 | | BRAFV600E Kinase Domain In Complex with Chemically Linked Vemurafenib Inhibitor VEM-BISAMIDE | | Descriptor: | 2,2'-oxybis(N-{[4-(3-{2,6-difluoro-3-[(propane-1-sulfonyl)amino]benzoyl}-1H-pyrrolo[2,3-b]pyridin-5-yl)phenyl]methyl}acetamide), BENZAMIDINE, Serine/threonine-protein kinase B-raf | | Authors: | Grasso, M.J, Marmorstein, R. | | Deposit date: | 2016-05-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Chemically Linked Vemurafenib Inhibitors Promote an Inactive BRAF(V600E) Conformation.
Acs Chem.Biol., 11, 2016
|
|
1DUX
 
 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
2IV0
 
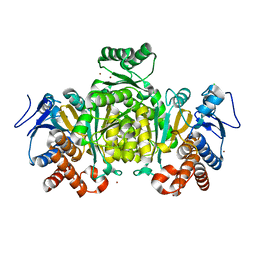 | | Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure analysis and engineering of chimers | | Descriptor: | CHLORIDE ION, ISOCITRATE DEHYDROGENASE, ZINC ION | | Authors: | Stokke, R, Karlstrom, M, Yang, N, Leiros, I, Ladenstein, R, Birkeland, N.K, Steen, I.H. | | Deposit date: | 2006-06-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thermal Stability of Isocitrate Dehydrogenase from Archaeoglobus Fulgidus Studied by Crystal Structure Analysis and Engineering of Chimers
Extremophiles, 11, 2007
|
|
6UV5
 
 | |
6UUW
 
 | | Structure of human ATP citrate lyase E599Q mutant in complex with Mg2+, citrate, ATP and CoA | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-11-01 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase
Nat.Struct.Mol.Biol., 27, 2020
|
|
7RB3
 
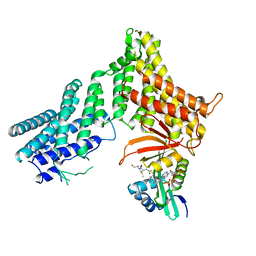 | |
1EJB
 
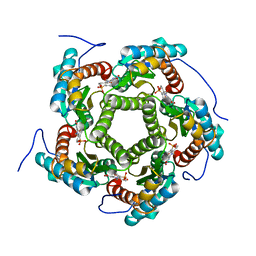 | | LUMAZINE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4-DIHYDROXYPYRIMIDIN-5-YL)-1-PENTYL-PHOSPHONIC ACID, LUMAZINE SYNTHASE | | Authors: | Meining, W, Mortl, S, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue.
J.Mol.Biol., 299, 2000
|
|
7RIG
 
 | | Structure of ACLY-D1026A-substrates | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-19 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7RKZ
 
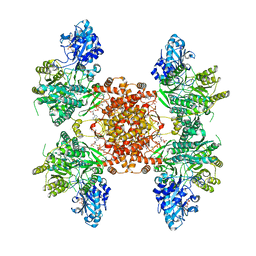 | | Structure of ACLY D1026A-substrates-asym-int | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-22 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7RMP
 
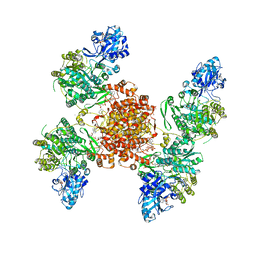 | | Structure of ACLY D1026A - substrates-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
1SBY
 
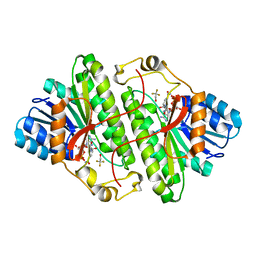 | | Alcohol dehydrogenase from Drosophila lebanonensis complexed with NAD+ and 2,2,2-trifluoroethanol at 1.1 A resolution | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL | | Authors: | Benach, J, Meijers, R, Atrian, S, Gonzalez-Duarte, R, Lamzin, V.S, Ladenstein, R. | | Deposit date: | 2004-02-11 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1-A crystal structure of D. lebanonensis ADH complexed with NAD+ and 2,2,2-trifluoroethanol
To be Published
|
|
6UI9
 
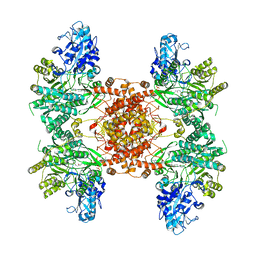 | |
3RJ5
 
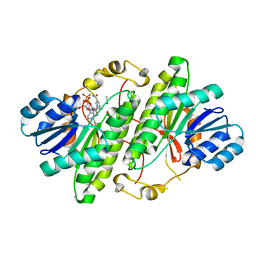 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
6VP9
 
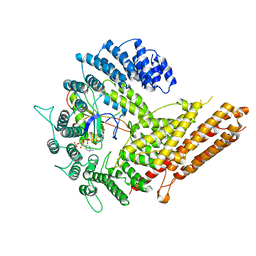 | | Cryo-EM structure of human NatB complex | | Descriptor: | CARBOXYMETHYL COENZYME *A, MDVFM peptide, N-alpha-acetyltransferase 20, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for N-terminal alpha-synuclein acetylation by human NatB.
Elife, 9, 2020
|
|
3RJ9
 
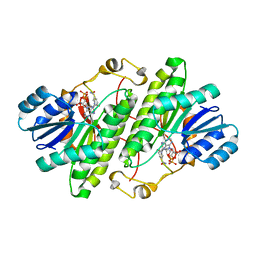 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
5WI2
 
 | | Crystal structure of the KA1 domain from human Chk1 | | Descriptor: | ACETATE ION, GLYCEROL, cDNA FLJ56409, ... | | Authors: | Emptage, R.P, Marmorstein, R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Intramolecular autoinhibition of checkpoint kinase 1 is mediated by conserved basic motifs of the C-terminal kinase-associated 1 domain.
J. Biol. Chem., 292, 2017
|
|
1B14
 
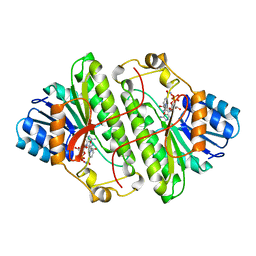 | | Alcohol Dehydrogenase from Drosophila Lebanonensis Binary Complex with NAD+ | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-25 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
9B3T
 
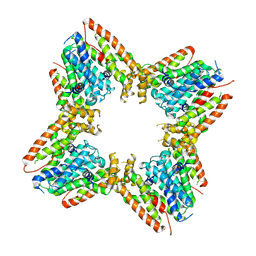 | |
9AYM
 
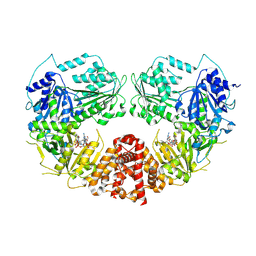 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-acetone-CoA bisubstrate probe | | Descriptor: | RNA cytidine acetyltransferase, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-[2-[3-[1-[(2~{R},3~{R},4~{R},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]sulfanyl-2-oxidanylidene-propyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
9B0I
 
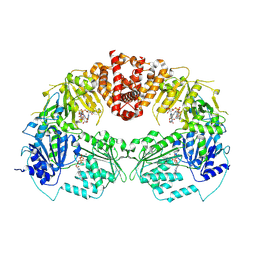 | |
9B0E
 
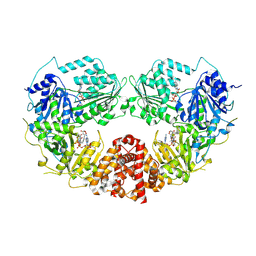 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-amide-CoA bisubstrate probe and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA cytidine acetyltransferase, [[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[[1-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]amino]-2-oxidanylidene-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
4R5K
 
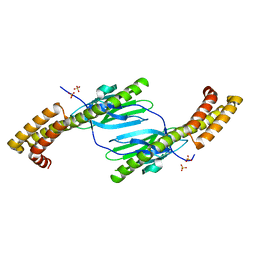 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-B) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, SULFATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7469 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5J
 
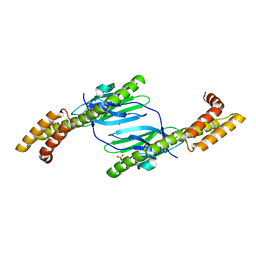 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-A) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
