8SF2
 
 | | Crystal structure of the engineered SsoPox variant IG7 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SF9
 
 | | Crystal structure of the engineered SsoPox variant IG7 - Alternative state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFC
 
 | | Crystal structure of the engineered SsoPox variant IVA4 in alternate state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFK
 
 | | Crystal structure of the engineered SsoPox variant IVE2 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFD
 
 | | Crystal structure of the engineered SsoPox variant IVB10 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
5NFF
 
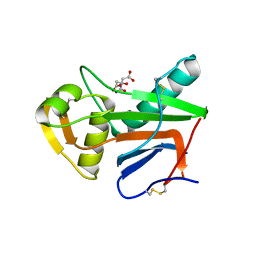 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
7Y13
 
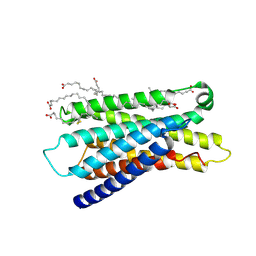 | | Cryo-EM structure of apo-state MrgD-Gi complex (local) | | Descriptor: | PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y12
 
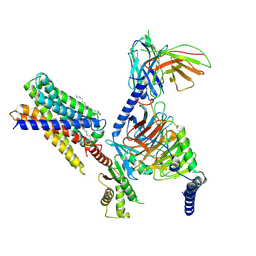 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine | | Descriptor: | BETA-ALANINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y14
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine (local) | | Descriptor: | BETA-ALANINE, PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y15
 
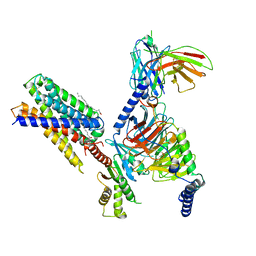 | | Cryo-EM structure of apo-state MrgD-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
6K8Q
 
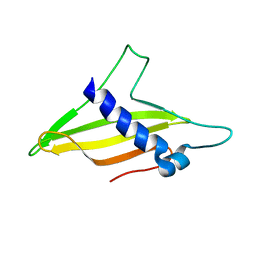 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
1BFG
 
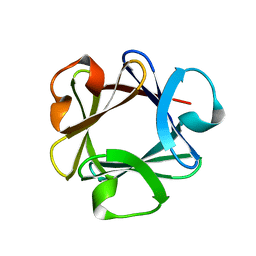 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
7W9Q
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-14 | | Descriptor: | (2~{R})-2-(3-chloranyl-4-oxidanyl-phenyl)-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, CALCIUM ION, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
7W9R
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-18 | | Descriptor: | (2~{R})-2-[3,5-bis(chloranyl)-4-oxidanyl-phenyl]-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
6K7F
 
 | | Crystal structure of MBPholo-Tim21 fusion protein with a 17-residue helical linker | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Mitochondrial import inner membrane translocase subunit TIM21, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bala, S, Shimada, A, Kohda, D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6K7D
 
 | |
6K7E
 
 | |
2PT5
 
 | | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Shikimate kinase | | Authors: | Jeyakanthan, J, Nithya, N, Shimada, A, Velmurugan, D, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5
To be Published
|
|
1AO6
 
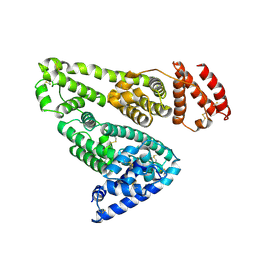 | |
1BM0
 
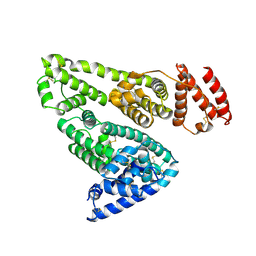 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN | | Descriptor: | SERUM ALBUMIN | | Authors: | Sugio, S, Kashima, A, Mochizuki, S, Noda, M, Kobayashi, K. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin at 2.5 A resolution.
Protein Eng., 12, 1999
|
|
1IVS
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS VALYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(VAL) AND VALYL-ADENYLATE ANALOGUE | | Descriptor: | N-[VALINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, Valyl-tRNA synthetase, tRNA (Val) | | Authors: | Fukai, S, Nureki, O, Sekine, S.-I, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-29 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
6J8M
 
 | | Low-dose structure of bovine heart cytochrome c oxidase in the fully oxidized state determined using 30 keV X-ray | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ueno, G, Shimada, A, Yamashita, E, Hasegawa, K, Kumasaka, T, Shinzawa-Itoh, K, Yoshikawa, S, Tsukihara, T, Yamamoto, M. | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-dose X-ray structure analysis of cytochrome c oxidase utilizing high-energy X-rays.
J.Synchrotron Radiat., 26, 2019
|
|
7F81
 
 | | Structure of the bacterial cellulose synthase subunit Z from Enterobacter sp. CJF-002 | | Descriptor: | GLYCEROL, Glucanase, S,R MESO-TARTARIC ACID | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
