2RRB
 
 | | Refinement of RNA binding domain in human Tra2 beta protein | | Descriptor: | cDNA FLJ40872 fis, clone TUTER2000283, highly similar to Homo sapiens transformer-2-beta (SFRS10) gene | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
3VYW
 
 | | Crystal structure of MNMC2 from Aquifex Aeolicus | | Descriptor: | BENZAMIDINE, MNMC2, S-ADENOSYLMETHIONINE | | Authors: | Shibata, R, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and structure of the Aquifex aeolicus protein DUF752: a bacterial tRNA-methyltransferase (MnmC2) functioning without the usually fused oxidase domain (MnmC1).
J.Biol.Chem., 287, 2012
|
|
2QQ1
 
 | | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5
To be Published
|
|
3SUV
 
 | |
3U6Y
 
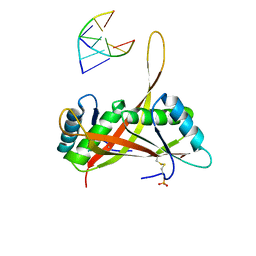 | |
3SUR
 
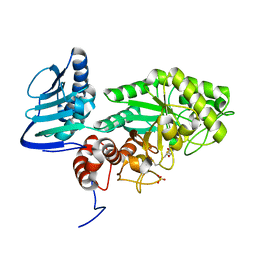 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with NAG-thiazoline. | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase, SULFATE ION | | Authors: | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-07-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
2ROZ
 
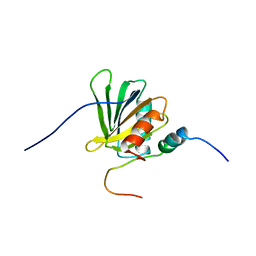 | | Structure of the C-terminal PID Domain of Fe65L1 Complexed with the Cytoplasmic Tail of APP Reveals a Novel Peptide Binding Mode | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2, peptide from Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2RPP
 
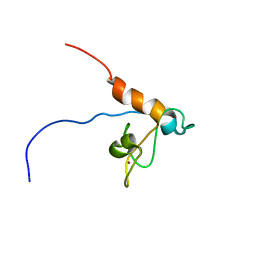 | | Solution structure of Tandem zinc finger domain 12 in Muscleblind-like protein 2 | | Descriptor: | Muscleblind-like protein 2, ZINC ION | | Authors: | Abe, C, Dang, W, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
3SUS
 
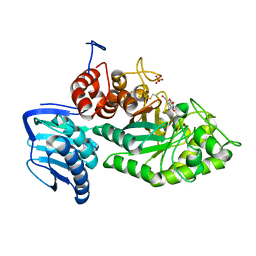 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-NAG-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, Beta-hexosaminidase, SULFATE ION | | Authors: | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-07-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
2RMX
 
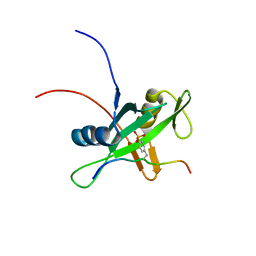 | | Solution structure of the SHP-1 C-terminal SH2 domain complexed with a tyrosine-phosphorylated peptide from NKG2A | | Descriptor: | NKG2-A/NKG2-B type II integral membrane protein, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Kasai, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of the two NKG2A immunoreceptor tyrosine-based inhibitory motifs (ITIMs) by the C-terminal SH2 domain of protein tyrosine phosphatase SHP-1
To be Published
|
|
3SUW
 
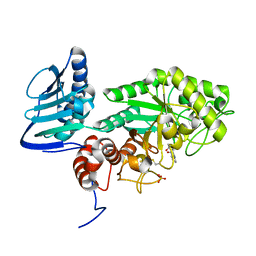 | |
2ROK
 
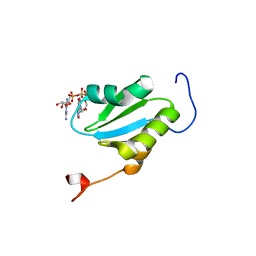 | | Solution structure of the cap-binding domain of PARN complexed with the cap analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, poly(A)-specific ribonuclease | | Authors: | Nagata, T, Suzuki, S, Endo, R, Shirouzu, M, Terada, T, Inoue, M, Kigawa, T, Guntert, P, Hayashizaki, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition.
Nucleic Acids Res., 36, 2008
|
|
3SUT
 
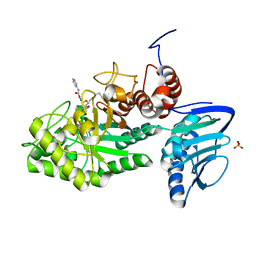 | |
3SUU
 
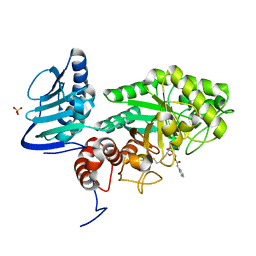 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-PUGNAc | | Descriptor: | Beta-hexosaminidase, SULFATE ION, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-07-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
2RPJ
 
 | | Solution structure of Fn14 CRD domain | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | He, F, Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain in Fn14, a member of the tumor necrosis factor receptor superfamily
Protein Sci., 18, 2009
|
|
2RS7
 
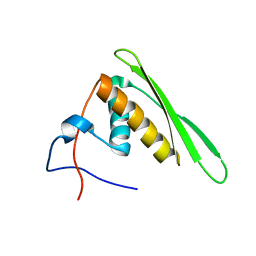 | | Solution structure of the second dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RUG
 
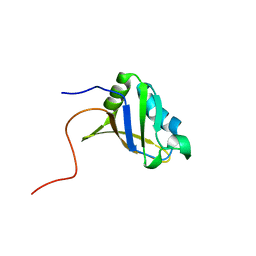 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
2QQ3
 
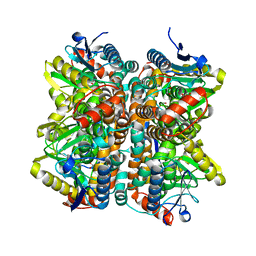 | | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase subunit I | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426
To be Published
|
|
2RS8
 
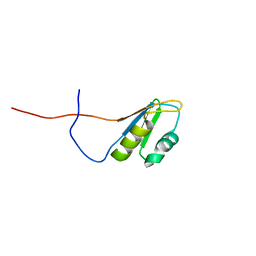 | | Solution structure of the N-terminal RNA recognition motif of NonO | | Descriptor: | Non-POU domain-containing octamer-binding protein | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA recognition motif of NonO
To be Published
|
|
3VIU
 
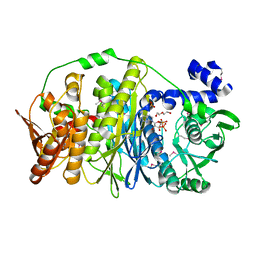 | | Crystal structure of PurL from thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Suzuki, S, Yanai, H, Kanagawa, M, Tamura, S, Watanabe, Y, Fuse, K, Baba, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-10-12 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2RNL
 
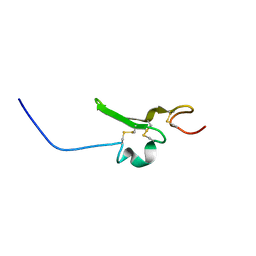 | | Solution structure of the EGF-like domain from human Amphiregulin | | Descriptor: | Amphiregulin | | Authors: | Qin, X, Hayashi, F, Terada, T, Shirouzu, M, Watanabe, S, Kigawa, T, Yabuta, N, Nojima, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the EGF-like domain from human Amphiregulin
To be Published
|
|
2ROR
 
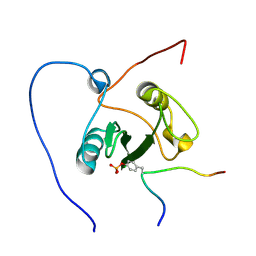 | | Solution structure of the VAV1 SH2 domain complexed with a tyrosine-phosphorylated peptide from SLP76 | | Descriptor: | 15-meric peptide from Lymphocyte cytosolic protein 2, Proto-oncogene vav | | Authors: | Tanaka, M, Kasai, T, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the VAV1 SH2 domain complexed with a tyrosine-phosphorylated peptide from SLP76
To be Published
|
|
2U2F
 
 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN OF HU2AF65 | | Descriptor: | PROTEIN (SPLICING FACTOR U2AF 65 KD SUBUNIT) | | Authors: | Ito, T, Muto, Y, Green, M.R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-05-26 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)).
EMBO J., 18, 1999
|
|
2RPZ
 
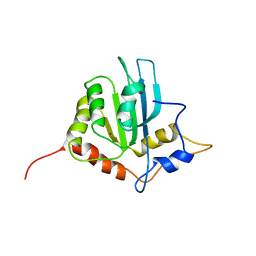 | | Solution structure of the monomeric form of mouse APOBEC2 | | Descriptor: | Probable C->U-editing enzyme APOBEC-2, ZINC ION | | Authors: | Hayashi, F, Nagata, T, Nagashima, T, Muto, Y, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-12-11 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monomeric form of mouse APOBEC2
To be Published
|
|
2RSF
 
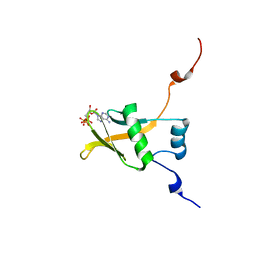 | | Complex structure of WWE in RNF146 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, E3 ubiquitin-protein ligase RNF146 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-01-31 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Complex structure of WWE domain in RNF146 with ATP
To be Published
|
|
