2B1F
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4 | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
1ZVA
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
3UIA
 
 | |
2B22
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4, SODIUM ION | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
1QR9
 
 | | INHIBITION OF HIV-1 INFECTIVITY BY THE GP41 CORE: ROLE OF A CONSERVED HYDROPHOBIC CAVITY IN MEMBRANE FUSION | | Descriptor: | GP41 ENVELOPE PROTEIN | | Authors: | Ji, H, Shu, W, Burling, F.T, Jiang, S.B, Lu, M. | | Deposit date: | 1999-06-18 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion.
J.Virol., 73, 1999
|
|
1QR8
 
 | | INHIBITION OF HIV-1 INFECTIVITY BY THE GP41 CORE: ROLE OF A CONSERVED HYDROPHOBIC CAVITY IN MEMBRANE FUSION | | Descriptor: | GP41 ENVELOPE PROTEIN | | Authors: | Ji, H, Shu, W, Burling, F.T, Jiang, S.B, Lu, M. | | Deposit date: | 1999-06-18 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion.
J.Virol., 73, 1999
|
|
2NRN
 
 | |
1DF5
 
 | |
1EQ7
 
 | |
1DF4
 
 | |
1DLB
 
 | | HELICAL INTERACTIONS IN THE HIV-1 GP41 CORE REVEALS STRUCTURAL BASIS FOR THE INHIBITORY ACTIVITY OF GP41 PEPTIDES | | Descriptor: | HIV-1 ENVELOPE GLYCOPROTEIN GP41 | | Authors: | Shu, W, Liu, J, Ji, H, Rading, L, Jiang, S, Lu, M. | | Deposit date: | 1999-12-09 | | Release date: | 1999-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides.
Biochemistry, 39, 2000
|
|
1S18
 
 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
4O8W
 
 | | Crystal Structure of the GerD spore germination protein | | Descriptor: | Spore germination protein | | Authors: | Li, Y, Jin, K, Ghosh, S, Devarakonda, P, Carlson, K, Davis, A, Stewart, K, Cammett, E, Rossi, P.P, Setlow, B, Lu, M, Setlow, P, Hao, B. | | Deposit date: | 2013-12-30 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Analysis of the GerD Spore Germination Protein of Bacillus Species.
J.Mol.Biol., 426, 2014
|
|
1S1D
 
 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
1Q47
 
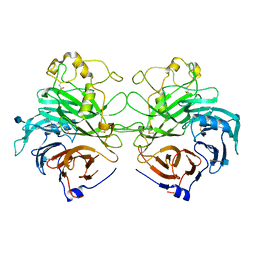 | | Structure of the Semaphorin 3A Receptor-Binding Module | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin 3A | | Authors: | Antipenko, A, Himanen, J.-P, van Leyen, K, Nardi-Dei, V, Lesniak, J, Barton, W.A, Rajashankar, K.R, Lu, M, Hoemme, C, Puschel, A, Nikolov, D. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the semaphorin-3A receptor binding module.
Neuron, 39, 2003
|
|
2R2V
 
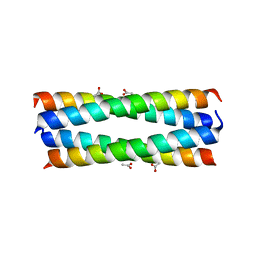 | |
2PFD
 
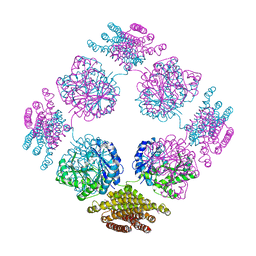 | | Anisotropically refined structure of FTCD | | Descriptor: | Formimidoyltransferase-cyclodeaminase | | Authors: | Poon, B.K, Chen, X, Lu, M, Quiocho, F.A, Wang, Q, Ma, J. | | Deposit date: | 2007-04-04 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Anisotropically refined structure of FTCD
To be Published
|
|
7SD4
 
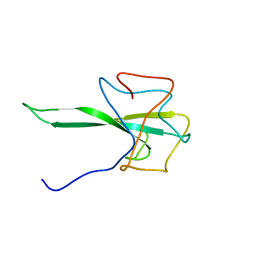 | | SARS-CoV-2 Nucleocapsid N-terminal domain (N-NTD) protein | | Descriptor: | Nucleoprotein | | Authors: | Sarkar, S, Runge, B, Russell, R.W, Calero, D, Zeinalilathori, S, Quinn, C.M, Lu, M, Calero, G, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Structure of SARS-CoV-2 Nucleocapsid Protein N-Terminal Domain.
J.Am.Chem.Soc., 144, 2022
|
|
4HLR
 
 | |
6J20
 
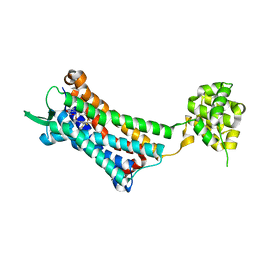 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
6J21
 
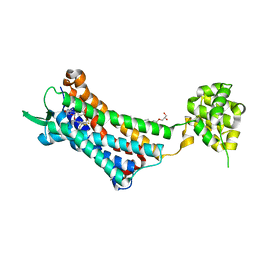 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
5ESW
 
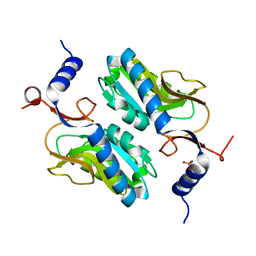 | | Crystal structure of Apo hypoxanthine-guanine phosphoribosyltransferase from Legionella pneumophila | | Descriptor: | PHOSPHATE ION, Purine/pyrimidine phosphoribosyltransferase | | Authors: | Zhang, N, Gong, X, Lu, M, Chen, X, Qin, X, Ge, H. | | Deposit date: | 2015-11-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Apo and GMP bound hypoxanthine-guanine phosphoribosyltransferase from Legionella pneumophila and the implications in gouty arthritis
J.Struct.Biol., 194, 2016
|
|
9JI7
 
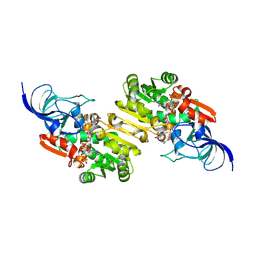 | | NADP-dependent oxidoreductase complexed with NADP and substrate 2 | | Descriptor: | (1Z,3E,5E,7S,8R,9S,10S,11R,13R,15S,16Z,18E,24S)-11-ethyl-2,7-dihydroxy-10-methyl-21,25-diazatetracyclo[22.2.1.08,15.09,13]heptacosa-1,3,5,16,18-pentaene-20,26,27-trione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase | | Authors: | Li, Y, Zhu, D, Xie, X, Li, F, Lu, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for Medium-Chain Dehydrogenase/Reductase-Catalyzed Reductive Cyclization in Polycyclic Tetramate Macrolactam Biosynthesis.
J.Am.Chem.Soc., 147, 2025
|
|
9JML
 
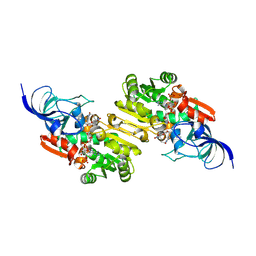 | | NADP-dependent oxidoreductase complexed with NADP and substrate 1 | | Descriptor: | (1Z,3Z,5Z,7S,8R,9S,10S,11R,13R,15R,16Z,18Z,24S)-11-ethyl-2,7-dihydroxy-10-methyl-21,25-diazatetracyclo[22.2.1.08,15.09,13]heptacosa-1,3,5,16,18-pentaene-20,26,27-trione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase | | Authors: | Li, Y, Zhu, D, Xie, X, Li, F, Lu, M. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Medium-Chain Dehydrogenase/Reductase-Catalyzed Reductive Cyclization in Polycyclic Tetramate Macrolactam Biosynthesis.
J.Am.Chem.Soc., 147, 2025
|
|
