5MNV
 
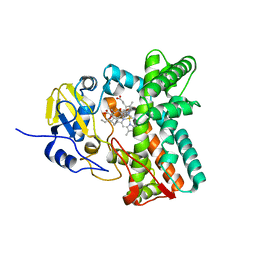 | |
5N9X
 
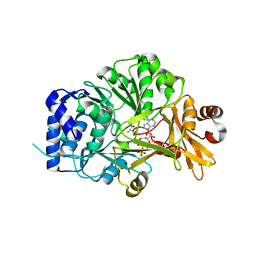 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, ligand bound structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylation domain, MAGNESIUM ION, ... | | Authors: | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
7JYZ
 
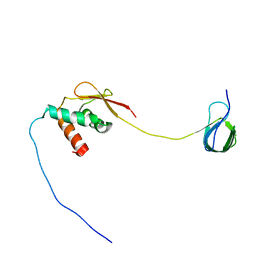 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
5N9W
 
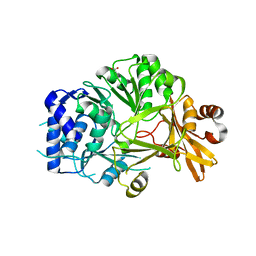 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, apo structure | | Descriptor: | ACETATE ION, Adenylation domain | | Authors: | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
5KHI
 
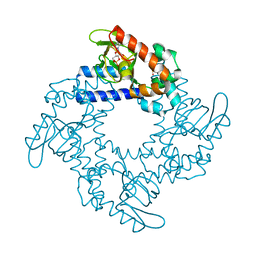 | | HCN2 CNBD in complex with purine riboside-3', 5'-cyclic monophosphate (cPuMP) | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Purine riboside-3',5'-cyclic monophosphate | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
5KHJ
 
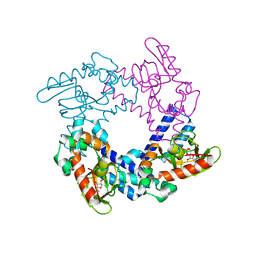 | | HCN2 CNBD in complex with uridine-3', 5'-cyclic monophosphate (cUMP) | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Uridine-3',5'-cyclic monophosphate | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
5KHK
 
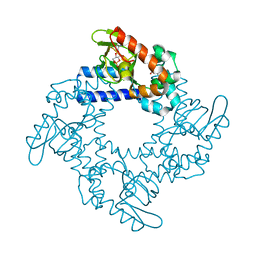 | | HCN2 CNBD in complex with 2-aminopurine riboside-3', 5'-cyclic monophosphate (2-NH2-cPuMP) | | Descriptor: | 2-Aminopurine riboside-3',5'-cyclic monophosphate, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
5KHG
 
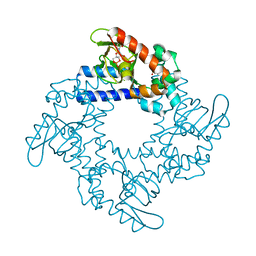 | | HCN2 CNBD in complex with cytidine-3', 5'-cyclic monophosphate (cCMP) | | Descriptor: | 4-amino-1-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxidotetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]pyrimidin-2(1H)-one, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
7M0E
 
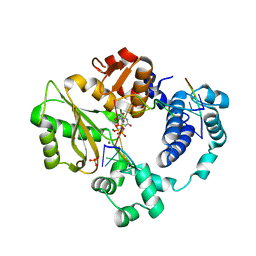 | | Pre-catalytic synaptic complex of DNA Polymerase Lambda with gapped DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M09
 
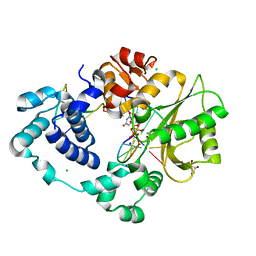 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with blunt-ended DSB substrate and incoming dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0A
 
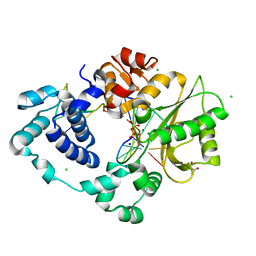 | | Incomplete in crystallo incorporation by DNA Polymerase Lambda bound to blunt-ended DSB substrate and incoming dTTP | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
5KHH
 
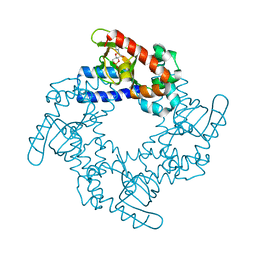 | | HCN2 CNBD in complex with inosine-3', 5'-cyclic monophosphate (cIMP) | | Descriptor: | Inosine-3',5'-cyclic monophosphate, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
7M0D
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound complementary DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0B
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound mismatched DSB and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0C
 
 | | Post-catalytic nicked complex of DNA Polymerase Lambda with bound mismatched DSB substrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Not Published
|
|
7M08
 
 | | Post-catalytic nicked complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Not Published
|
|
7M07
 
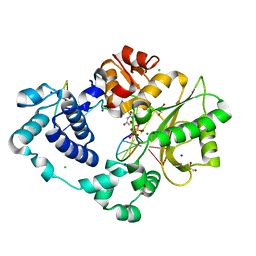 | | Pre-catalytic ternary complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
8A58
 
 | |
1FX1
 
 | |
6HK2
 
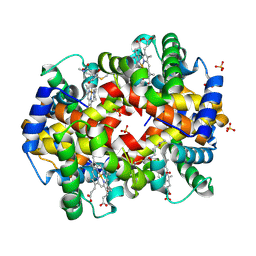 | | Crystal structure of ferric R-state human methemoglobin bound to maleimide-deferoxamine bifunctional chelator (DFO) | | Descriptor: | 3-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]-~{N}-methyl-propanamide, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Boffi, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biodistribution PET/CT Study of Hemoglobin-DFO-89Zr Complex in Healthy and Lung Tumor-Bearing Mice.
Int J Mol Sci, 21, 2020
|
|
6I3T
 
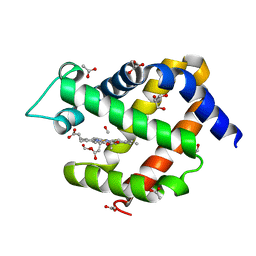 | | Crystal structure of murine neuroglobin bound to CO at 40 K. | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C. | | Deposit date: | 2018-11-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
6I40
 
 | | Crystal structure of murine neuroglobin bound to CO at 15K under illumination using optical fiber | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C, Vallone, B. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
6IDW
 
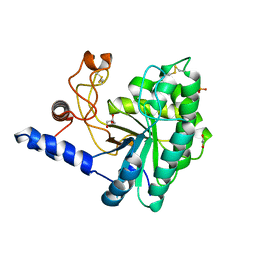 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2018-09-11 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structures of the GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo and endo activity and its complex with cellobiose.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6H09
 
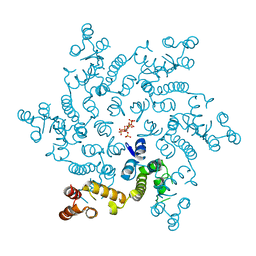 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2018-07-06 | | Release date: | 2018-08-15 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
6H6I
 
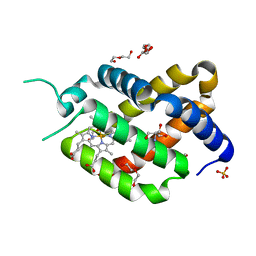 | | Ferric murine neuroglobin Gly-loop mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
