4O03
 
 | | Crystal structure of Ca2+ bound prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7E94
 
 | | Intact TRAPPII (State II) | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E2D
 
 | | Monomer of TRAPPII (Closed) | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Sui, S.F, Sun, S, Mi, C.C. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E8T
 
 | | Monomer of Ypt32-TRAPPII | | Descriptor: | GTP-binding protein YPT32/YPT11, TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-02 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E93
 
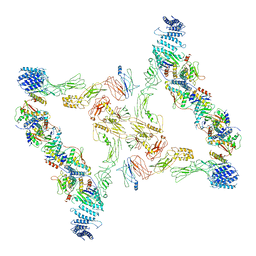 | | Intact TRAPPII (state III). | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.54 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E2C
 
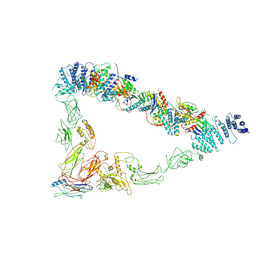 | | Monomer of TRAPPII (open) | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Sui, S.F, Sun, S, Mi, C.C. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7EA3
 
 | | Intact Ypt32-TRAPPII (dimer). | | Descriptor: | GTP-binding protein YPT32/YPT11, TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E8S
 
 | | Intact TRAPPII (state I). | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-02 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
5V88
 
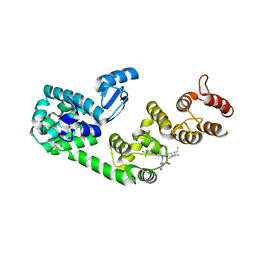 | | Structure of DCN1 bound to NAcM-COV | | Descriptor: | Lysozyme,DCN1-like protein 1, N-{2-[({1-[(2R)-pentan-2-yl]piperidin-4-yl}{[3-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]phenyl}propanamide | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5VJ0
 
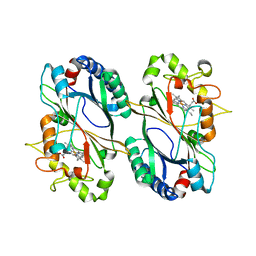 | |
5V89
 
 | | Structure of DCN4 PONY domain bound to CUL1 WHB | | Descriptor: | Cullin-1, DCN1-like protein 4 | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | Descriptor: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V83
 
 | | Structure of DCN1 bound to NAcM-HIT | | Descriptor: | Lysozyme,DCN1-like protein 1 chimera, N-(1-benzylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
7XQ8
 
 | | Structure of human B-cell antigen receptor of the IgM isotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Chen, M.Y, Su, Q, Shi, Y.G. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human IgM B cell receptor.
Science, 377, 2022
|
|
5D5Q
 
 | | HcgB from Methanocaldococcus jannaschii with the pyridinol derived from FeGP cofactor of [Fe]-hydrogenase | | Descriptor: | (4,6-dihydroxy-3,5-dimethylpyridin-2-yl)acetic acid, Uncharacterized protein MJ0488, Guanylyltransferase | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Towards artificial methanogenesis: biosynthesis of the [Fe]-hydrogenase cofactor and characterization of the semi-synthetic hydrogenase.
Faraday Discuss., 198, 2017
|
|
5D5P
 
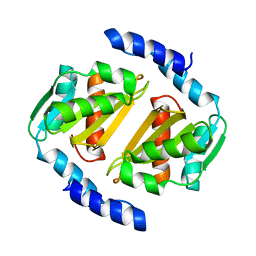 | | HcgB from Methanococcus maripaludis | | Descriptor: | HcgB | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards artificial methanogenesis: biosynthesis of the [Fe]-hydrogenase cofactor and characterization of the semi-synthetic hydrogenase.
Faraday Discuss., 198, 2017
|
|
7VZG
 
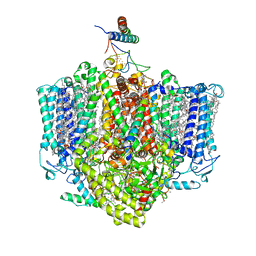 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
7VZR
 
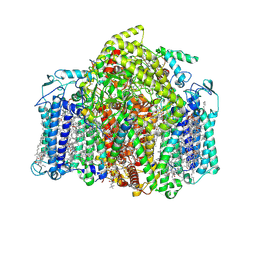 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
7WO9
 
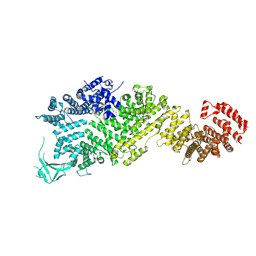 | | Cryo-EM structure of full-length Nup188 | | Descriptor: | Nucleoporin NUP188 | | Authors: | Zhao, L, Li, Z.Q, Sui, S.F. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOO
 
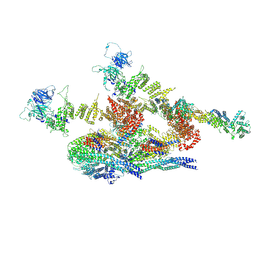 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOT
 
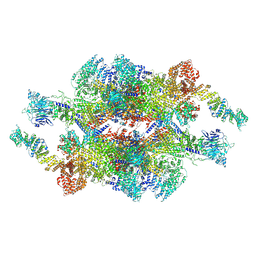 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
