8WMD
 
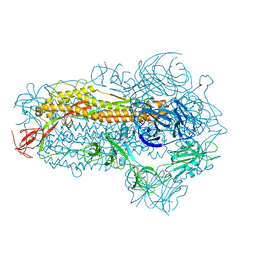 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
8K3D
 
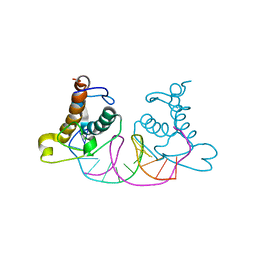 | | Crystal structure of NRF1 DBD bound to DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*CP*GP*CP*AP*TP*GP*CP*GP*CP*AP*CP*C)-3'), Nuclear respiratory factor 1 | | Authors: | Li, W.F, Liu, K, Min, J.R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of specific DNA sequence recognition by NRF1.
Nucleic Acids Res., 52, 2024
|
|
8K4L
 
 | |
2FZN
 
 | |
5NX7
 
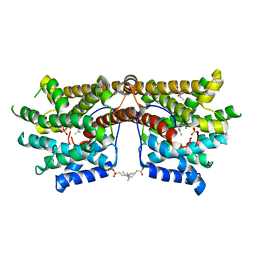 | | Crystal structure of 1,8-cineole synthase from Streptomyces clavuligerus in complex with 2-fluoroneryl diphosphate and 2-fluorogeranyl diphosphate | | Descriptor: | (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
5NX4
 
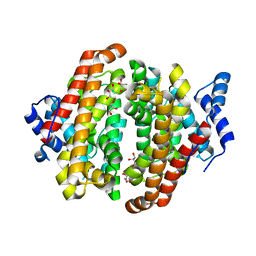 | |
8XLM
 
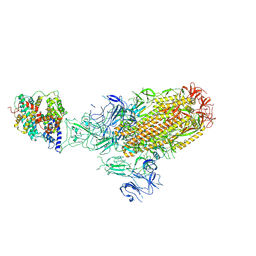 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
2FZM
 
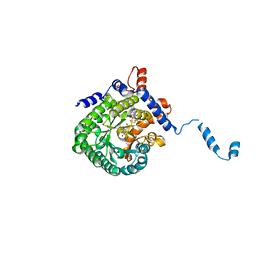 | |
8XLN
 
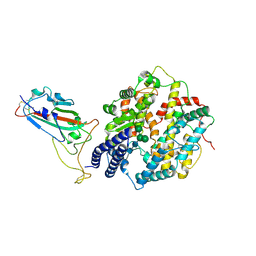 | | Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
6XPC
 
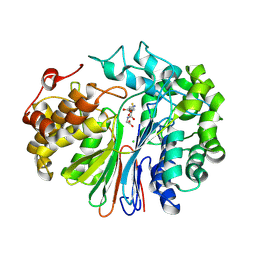 | | Structure of human GGT1 in complex with full GSH molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of glutathione- and inhibitor-bound human GGT1: critical interactions within the cysteinylglycine binding site.
J.Biol.Chem., 296, 2020
|
|
6U63
 
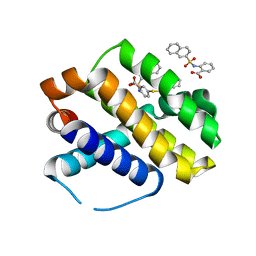 | | Mcl-1 bound to compound 17 | | Descriptor: | 2-{[(naphthalen-2-yl)sulfonyl]amino}-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
5NX6
 
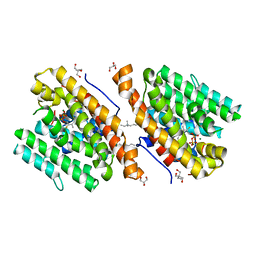 | | Crystal structure of 1,8-cineole synthase from Streptomyces clavuligerus in complex with 2-fluoroneryl diphosphate | | Descriptor: | (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
8A63
 
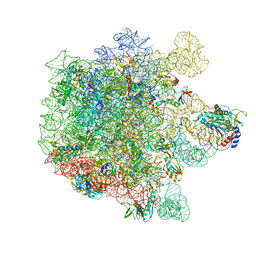 | | Cryo-EM structure of Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
8A5I
 
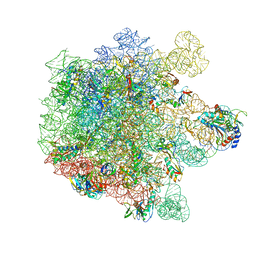 | | Cryo-EM structure of Lincomycin bound to the Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
8A57
 
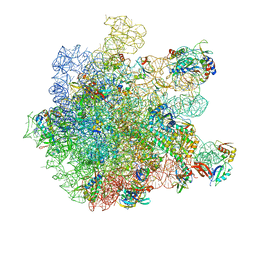 | | Cryo-EM structure of HflXr bound to the Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
6OE7
 
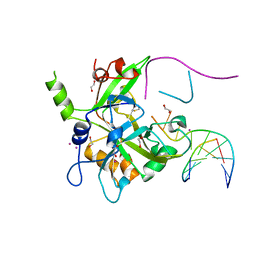 | | Crystal structure of HMCES cross-linked to DNA abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*(DRZ)P*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6U6F
 
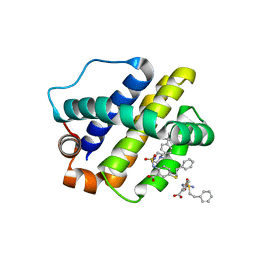 | | The crystal structure of anti-apoptotic Mcl-1 protein in complex with 2, 5-substituted benzoic acid inhibitor 21 | | Descriptor: | 2-[({4-[(4-tert-butylphenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Yang, Y, Stuckey, J.A, Nikolovska-Coleska, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
5NX5
 
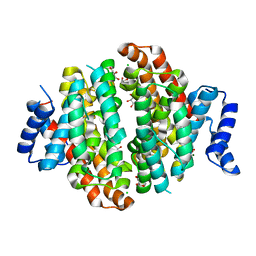 | | Crystal structure of Linalool/Nerolidol synthase from Streptomyces clavuligerus in complex with 2-fluorogeranyl diphosphate | | Descriptor: | (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
6UCM
 
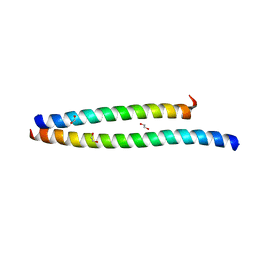 | | Transcription factor DeltaFosB bZIP domain self-assembly, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6A58
 
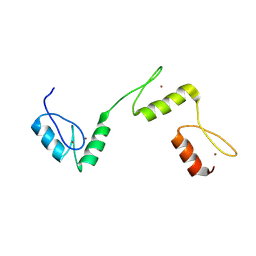 | | Structure of histone demethylase REF6 | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
5LJM
 
 | | Structure of SPATA2 PUB domain | | Descriptor: | GLYCEROL, Spermatogenesis-associated protein 2 | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
6WX5
 
 | | Adducts formed after 3 weeks in the reaction of chlorido[chlorido(2,2'-((2-([2,2':6',2''-Terpyridin]-4'-yloxy)ethyl)azanediyl)bis(ethan-1-ol))platinum(II)] with HEWL | | Descriptor: | ACETATE ION, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mustards-Derived Terpyridine-Platinum Complexes as Anticancer Agents: DNA Alkylation vs Coordination.
Inorg.Chem., 60, 2021
|
|
6A57
 
 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
8IVW
 
 | | Crystal structure of NRP2 in complex with aNRP2-10 Fab fragment | | Descriptor: | Heavy chian of antibody 10V8 Fab fragment, Light chain of antibody 10V8 Fab fragment, Neuropilin-2 | | Authors: | Geng, Y, Zhai, L. | | Deposit date: | 2023-03-29 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer.
Sci Transl Med, 15, 2023
|
|
8IVX
 
 | | Crystal structure of NRP2 in complex with aNRP2-14 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody 14V4 Fab fragment, Light chain of antibody 14V4 Fab fragment, ... | | Authors: | Geng, Y, Zhai, L. | | Deposit date: | 2023-03-29 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer.
Sci Transl Med, 15, 2023
|
|
