4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LWV
 
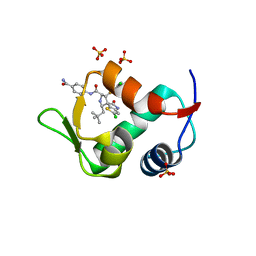 | | The 2.3A Crystal Structure of Humanized Xenopus MDM2 with RO5545353 | | Descriptor: | (2S,3R,4R,5R)-N-(4-carbamoyl-2-methoxyphenyl)-2'-chloro-4-(3-chloro-2-fluorophenyl)-2-(2,2-dimethylpropyl)-5'-oxo-4',5'-dihydrospiro[pyrrolidine-3,6'-thieno[3,2-b]pyrrole]-5-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Potent and Orally Active p53-MDM2 Inhibitors RO5353 and RO2468 for Potential Clinical Development.
ACS MED.CHEM.LETT., 5, 2014
|
|
6EBA
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Unoccupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-08-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
8U8L
 
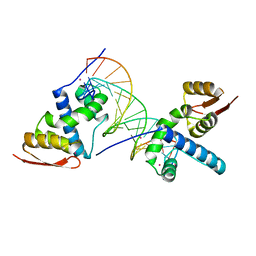 | | X-ray crystal structure of TEBP-2 MCD3 with ds DNA | | Descriptor: | CESIUM ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*CP*TP*TP*AP*GP*GP*CP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U8M
 
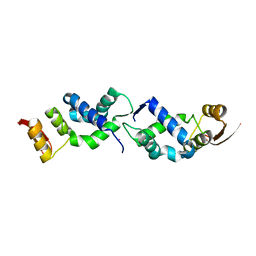 | | X-ray crystal structure of TEBP-1 MCD2 homodimer | | Descriptor: | COBALT (II) ION, Double-strand telomeric DNA-binding proteins 1 | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6E9C
 
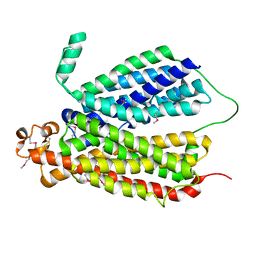 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
4LWT
 
 | | The 1.6A Crystal Structure of Humanized Xenopus MDM2 with RO5027344 | | Descriptor: | (3S)-3-[(3R)-1-acetylpiperidin-3-yl]-6-chloro-3-(3-chlorobenzyl)-1,3-dihydro-2H-indol-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Kammlott, U. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
6L8T
 
 | |
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.35534 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14182 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
4RI2
 
 | | Crystal structure of the photoprotective protein PsbS from spinach | | Descriptor: | CHLOROPHYLL A, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RI3
 
 | | Crystal structure of DCCD-modified PsbS from spinach | | Descriptor: | DICYCLOHEXYLUREA, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8YLB
 
 | | Cocrystal structures of agonists compound 1 with HsClpP | | Descriptor: | 5-[(2-methylphenyl)methyl]-11-(phenylmethyl)-2,5,7,11-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),6-dien-8-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Zhao, N, Zhu, Y, Bao, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational Design of a Novel Class of Human ClpP Agonists through a Ring-Opening Strategy with Enhanced Antileukemia Activity.
J.Med.Chem., 67, 2024
|
|
8WTW
 
 | | Cryo-EM structure of noradrenaline transporter in complex with a x-MrlA analogue | | Descriptor: | CHLORIDE ION, MrlA, SODIUM ION, ... | | Authors: | Zhao, Y, Hu, T, Yu, Z. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Transport and inhibition mechanisms of the human noradrenaline transporter.
Nature, 632, 2024
|
|
8WTU
 
 | | Cryo-EM structure of noradrenaline transporter in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium-dependent noradrenaline transporter | | Authors: | Zhao, Y, Hu, T, Yu, Z. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Transport and inhibition mechanisms of the human noradrenaline transporter.
Nature, 632, 2024
|
|
8WTV
 
 | | Cryo-EM structure of noradrenaline transporter in complex with noradrenaline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Noradrenaline, ... | | Authors: | Zhao, Y, Hu, T, Yu, Z. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Transport and inhibition mechanisms of the human noradrenaline transporter.
Nature, 632, 2024
|
|
8WTY
 
 | | Cryo-EM structure of noradrenaline transporter in complex with ziprasidone | | Descriptor: | 5-[2-[4-(1,2-benzothiazol-3-yl)piperazin-1-yl]ethyl]-6-chloranyl-1,3-dihydroindol-2-one, Sodium-dependent noradrenaline transporter | | Authors: | Zhao, Y, Hu, T, Yu, Z. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transport and inhibition mechanisms of the human noradrenaline transporter.
Nature, 632, 2024
|
|
