1WPB
 
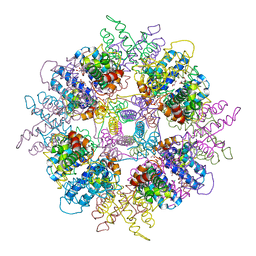 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
3JYF
 
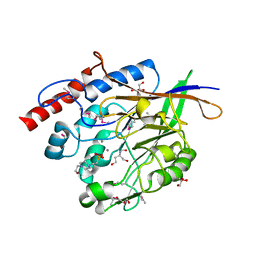 | | The crystal structure of a 2,3-cyclic nucleotide 2-phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor protein from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fan, Y, Volkart, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-21 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a 2,3-cyclic nucleotide 2-phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor protein from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
3QVQ
 
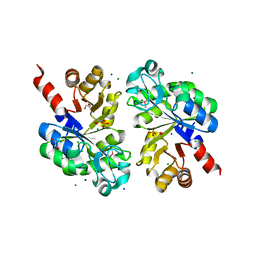 | | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Cuff, M.E, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate
To be Published
|
|
3RHF
 
 | | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Feldmann, B, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1
TO BE PUBLISHED
|
|
3OGH
 
 | | Crystal structure of yciE protein from E. coli CFT073, a member of ferritine-like superfamily of diiron-containing four-helix-bundle proteins | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Nocek, B, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of yciE protein from E. coli CFT073, a member of ferritine-like superfamily of diiron-containing four-helix-bundle proteins
To be Published
|
|
3OHR
 
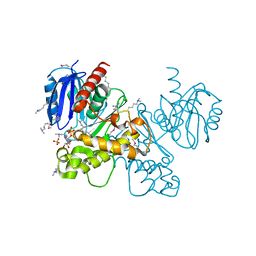 | | Crystal structure of fructokinase from bacillus subtilis complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative fructokinase, SULFATE ION, ... | | Authors: | Nocek, B, Volkart, L, Cuff, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
3OLQ
 
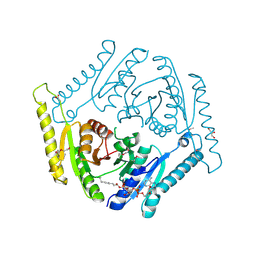 | | The crystal structure of a universal stress protein E from Proteus mirabilis HI4320 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | The crystal structure of a universal stress protein E from
Proteus mirabilis HI4320
To be Published
|
|
3OMT
 
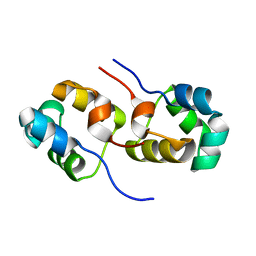 | | Putative antitoxin component, CHU_2935 protein, from Xre family from Prevotella buccae. | | Descriptor: | PHOSPHATE ION, uncharacterized protein | | Authors: | Osipiuk, J, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of putative antitoxin component, CHU_2935 protein, from Xre family from Prevotella buccae.
To be Published
|
|
1XIZ
 
 | | Structural Genomics, The crystal structure of domain IIA of putative phosphotransferase system specific for mannitol/fructose from Salmonella typhimurium | | Descriptor: | putative phosphotransferase system mannitol/fructose-specific IIA domain | | Authors: | Zhang, R, Joachimiak, G, Otwinowski, Z, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of domain IIA of putative phosphotransferase system specific for mannitol/fructose from Salmonella typhimurium
To be Published
|
|
3RQB
 
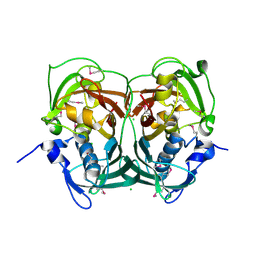 | |
3ROB
 
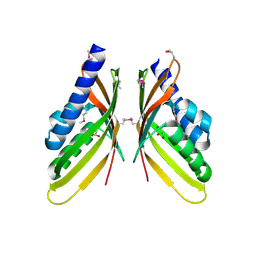 | |
3VDH
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 | | Descriptor: | B-1,4-endoglucanase, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
3RQ1
 
 | | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula | | Descriptor: | 2-OXOGLUTARIC ACID, Aminotransferase class I and II, CHLORIDE ION, ... | | Authors: | Kim, Y, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula
To be Published
|
|
2KKY
 
 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
3GEM
 
 | | Crystal structure of short-chain dehydrogenase from Pseudomonas syringae | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Nocek, B, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of short-chain dehydrogenase from Pseudomonas syringae.
To be Published
|
|
3D8U
 
 | |
3TOV
 
 | |
3NSX
 
 | | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174
To be Published
|
|
3O5V
 
 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
3GYG
 
 | | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis | | Descriptor: | MAGNESIUM ION, NTD biosynthesis operon putative hydrolase ntdB | | Authors: | Nocek, B, Stein, A, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis
To be Published
|
|
3TT2
 
 | | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GCN5-related N-acetyltransferase, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus
To be Published
|
|
3TY7
 
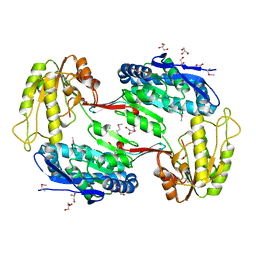 | | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Joachimiak, G, Jedrzejczak, R, Rubin, E, Ioerger, T, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus
To be Published
|
|
1U9C
 
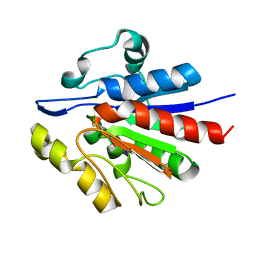 | | Crystallographic structure of APC35852 | | Descriptor: | APC35852 | | Authors: | Borek, D, Chen, Y, Shao, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of DJI superfamily
To be Published
|
|
3SRX
 
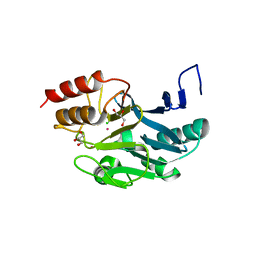 | | New Delhi Metallo-beta-Lactamase-1 Complexed with Cd | | Descriptor: | Beta-lactamase NDM-1, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 Complexed with Cd
To be Published
|
|
3NUQ
 
 | | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, A, Yang, C, Singer, A.U, Evdokimova, E, Kudritsdka, M, Brown, G, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae
To be Published
|
|
