4AHQ
 
 | | Crystal Structure of N-acetylneuraminic acid lyase mutant K165C from Staphylococcus aureus | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
2FD5
 
 | | The crystal structure of a transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | transcriptional regulator | | Authors: | Zhang, R, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
4IGS
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OHX
 
 | | C. Elegans Clp1 bound to ADP and Mg2+ (RNA released state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
2B0C
 
 | | The crystal structure of the putative phosphatase from Escherichia coli | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, putative phosphatase | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-13 | | Release date: | 2005-11-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A crystal structure of the putative phosphatase from Escherichia coli
To be Published
|
|
3LVM
 
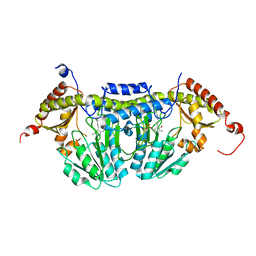 | | Crystal Structure of E.coli IscS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
4AHO
 
 | | Crystal Structure of N-acetylneuraminic acid lyase from Staphylococcus aureus with the chemical modification thia-lysine at position 165 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
2B1Y
 
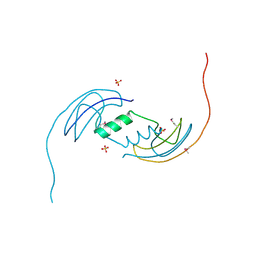 | | Crystal Structure of Protein of Unknown Function ATU1913 from Agrobacterium tumefaciens str. C58 | | Descriptor: | SULFATE ION, hypothetical protein Atu1913 | | Authors: | Nocek, B, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Hypothetical Protein from Agrobacterium tumefaciens
reveals a new fold.
To be Published
|
|
2B25
 
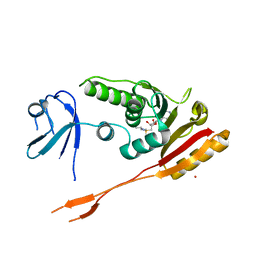 | | Human putative tRNA(1-methyladenosine)methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Tempel, W, Wu, H, Dong, A, Zeng, H, Loppnau, P, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-16 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human putative tRNA(1-methyladenosine)methyltransferase
To be Published
|
|
3LXQ
 
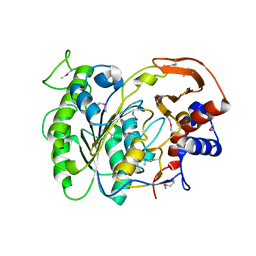 | | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A | | Descriptor: | CHLORIDE ION, Uncharacterized protein VP1736 | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A
To be Published
|
|
3LRD
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, Major ampullate spidroin 1, TRIETHYLENE GLYCOL | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Rising, A, Johansson, J, Knight, S.D, Saenz, A, Casals, C. | | Deposit date: | 2010-02-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
2B3M
 
 | | Crystal structure of protein AF1124 from Archaeoglobus fulgidus | | Descriptor: | hypothetical protein AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of protein AF1124 from Archaeoglobus fulgidus
To be Published
|
|
3LTJ
 
 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | AlphaRep-4 | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
4ASX
 
 | | Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with dihydro-Bauerine C | | Descriptor: | 1,2-ETHANEDIOL, 7,8-bis(chloranyl)-9-methyl-3,4-dihydro-2H-pyrido[3,4-b]indol-1-one, ACTIVIN RECEPTOR TYPE-2A | | Authors: | Williams, E, Chaikuad, A, Canning, P, Kochan, G, Mahajan, P, Cooper, C.D.O, Beltrami, A, Krojer, T, Pohl, B, Bracher, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A. | | Deposit date: | 2012-05-03 | | Release date: | 2012-05-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Activin Receptor Type-Iia (Acvr2A) Kinase Domain in Complex with a Beta- Carboline Inhibitor
To be Published
|
|
3LRG
 
 | | Structure of anti-huntingtin VL domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IMIDAZOLE, anti-huntingtin VL domain | | Authors: | Schiefner, A, Chatwell, L, Skerra, A. | | Deposit date: | 2010-02-11 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Disulfide-Free Single-Domain V(L) Intrabody with Blocking Activity towards Huntingtin Reveals a Novel Mode of Epitope Recognition.
J.Mol.Biol., 414, 2011
|
|
2B97
 
 | | Ultra-high resolution structure of hydrophobin HFBII | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Linder, M, Popov, A, Schmidt, A, Rouvinen, J. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4OVB
 
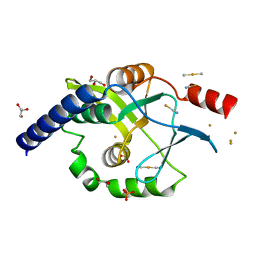 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor, Gold (I) Cyanide derivative | | Descriptor: | GLYCEROL, GOLD (I) CYANIDE ION, PHOSPHATE ION, ... | | Authors: | Maindola, P, Goyal, P, Arulandu, A. | | Deposit date: | 2014-02-21 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
1BXY
 
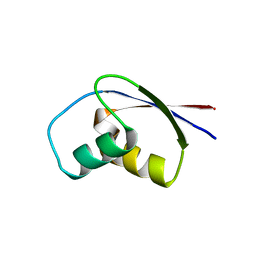 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1SYI
 
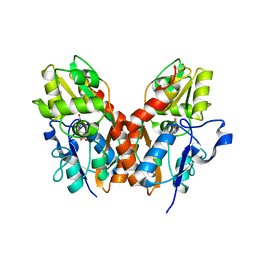 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
2B0V
 
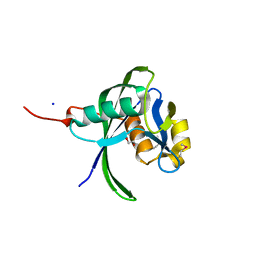 | | NUDIX hydrolase from Nitrosomonas europaea. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NUDIX hydrolase, ... | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
4JJD
 
 | |
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
1IBH
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT M41I | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
3LY9
 
 | |
2GFQ
 
 | | Structure of Protein of Unknown Function PH0006 from Pyrococcus horikoshii | | Descriptor: | MAGNESIUM ION, SULFATE ION, UPF0204 protein PH0006 | | Authors: | Cuff, M.E, Skarina, T, Gorodichtchenskaia, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of hypothetical protein ph0006 from Pyrococcus horikoshii
To be Published
|
|
