8PAY
 
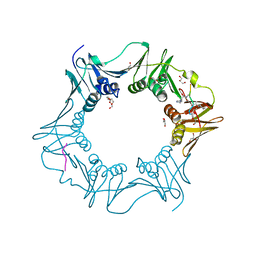 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | Descriptor: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
1TZ0
 
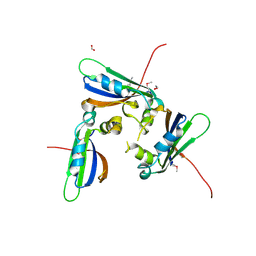 | |
6C9L
 
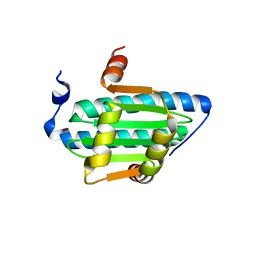 | | MEF2B Apo Protein Structure | | Descriptor: | Myocyte-specific enhancer factor 2B | | Authors: | Lei, X, Chen, L. | | Deposit date: | 2018-01-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Apo MEF2B Reveals New Insights in DNA Binding and Cofactor Interaction.
Biochemistry, 57, 2018
|
|
5DAY
 
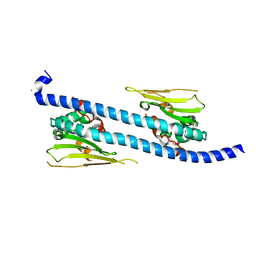 | | The structure of NAP1-Related Protein(NRP1) in Arabidopsis | | Descriptor: | CALCIUM ION, NAP1-related protein 1 | | Authors: | Zhu, Y, Rong, L, Yang, Y, Zhang, C, Feng, H.Y, Zheng, L.N, Shen, W.H, Ma, J.B, Dong, A.W. | | Deposit date: | 2015-08-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | The structure of NAP1-Related Protein(NRP1) in Arabidopsis
To Be Published
|
|
8PF4
 
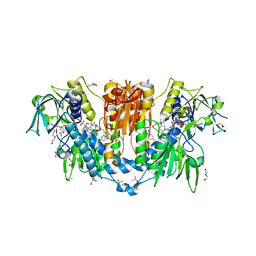 | | Crystal structure of Trypanosoma brucei trypanothione reductase in complex with 4-(((5-((4-fluorophenethyl)carbamoyl)furan-2-yl)methyl)(4-fluorophenyl)carbamoyl)-1-methyl-1-(3-phenylpropyl)piperazin-1-ium | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Exertier, C, Ilari, A, Fiorillo, A, Antonelli, L. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment Merging, Growing, and Linking Identify New Trypanothione Reductase Inhibitors for Leishmaniasis.
J.Med.Chem., 67, 2024
|
|
1U12
 
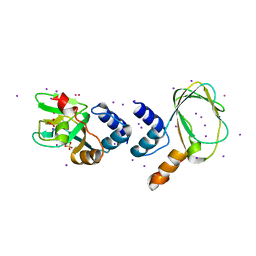 | | M. loti cyclic nucleotide binding domain mutant | | Descriptor: | IODIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Clayton, G.M, Silverman, W.R, Heginbotham, L, Morais-Cabral, J.H. | | Deposit date: | 2004-07-14 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Ligand Activation in a Cyclic Nucleotide Regulated Potassium Channel
Cell(Cambridge,Mass.), 119, 2004
|
|
1U6M
 
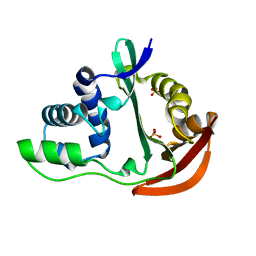 | | The crystal structure of acetyltransferase | | Descriptor: | SULFATE ION, acetyltransferase, GNAT family | | Authors: | Min, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of acetyltransferase, GNAT family from Enterococcus faecalis
To be Published
|
|
1U7D
 
 | |
1U9U
 
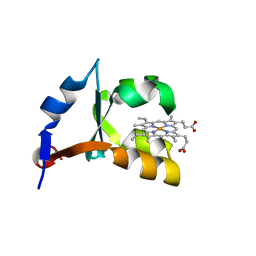 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8PAT
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
5CS0
 
 | |
5CSK
 
 | |
1VYH
 
 | | PAF-AH Holoenzyme: Lis1/Alfa2 | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB ALPHA SUBUNIT, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB BETA SUBUNIT | | Authors: | Tarricone, C, Perrina, F, Monzani, S, Massimiliano, L, Knapp, S, Tsai, L.-H, Derewenda, Z.S, Musacchio, A. | | Deposit date: | 2004-04-30 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Coupling Paf Signaling to Dynein Regulation: Structure of Lis1 in Complex with Paf-Acetylhydrolase.
Neuron, 44, 2004
|
|
5DVY
 
 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
1U6T
 
 | | Crystal structure of the human SH3 binding glutamic-rich protein like | | Descriptor: | CITRIC ACID, SH3 domain-binding glutamic acid-rich-like protein | | Authors: | Yin, L, Xiang, Y, Yang, N, Zhu, D.-Y, Huang, R.-H, Wang, D.-C. | | Deposit date: | 2004-08-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human SH3BGRL protein: the first structure of the human SH3BGR family representing a novel class of thioredoxin fold proteins
Proteins, 61, 2005
|
|
1W45
 
 | | The 2.5 Angstroem structure of the K16A mutant of annexin A8, which has an intact N-terminus. | | Descriptor: | ANNEXIN A8 | | Authors: | Rety, S, Sopkova-de Oliveira Santos, J, Raguenes-Nicol, C, Dreyfuss, L, Blondeau, K, Hofbauerova, K, Renouard, M, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The Crystal Structure of Annexin A8 is Similar to that of Annexin A3
J.Mol.Biol., 345, 2005
|
|
5E31
 
 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
8PF3
 
 | | Crystal structure of Trypanosoma brucei trypanothione reductase in complex with 1-(3,4-dichlorobenzyl)-4-(((5-((4-fluorophenethyl)carbamoyl)furan-2-yl)methyl)(4-fluorophenyl)carbamoyl)-1-(3-phenylpropyl)piperazin-1-ium | | Descriptor: | 4-[(3,4-dichlorophenyl)methyl]-~{N}-(4-fluorophenyl)-~{N}-[[5-[2-(4-fluorophenyl)ethylcarbamoyl]furan-2-yl]methyl]-4-(3-phenylpropyl)-1,4$l^{4}-diazinane-1-carboxamide, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Exertier, C, Ilari, A, Fiorillo, A, Antonelli, L. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment Merging, Growing, and Linking Identify New Trypanothione Reductase Inhibitors for Leishmaniasis.
J.Med.Chem., 67, 2024
|
|
5DZW
 
 | | Protocadherin alpha 4 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin alpha-4, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
1TVM
 
 | | NMR structure of enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system | | Descriptor: | PTS system, galactitol-specific IIB component | | Authors: | Volpon, L, Young, C.R, Lim, N.S, Iannuzzi, P, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system and its interaction with GatA.
Protein Sci., 15, 2006
|
|
5E23
 
 | | Human transthyretin (TTR) complexed with (2,7-Dibromo-fluoren-9-ylideneaminooxy)-acetic acid | | Descriptor: | DIMETHYL SULFOXIDE, Transthyretin, {[(2,7-dibromo-9H-fluoren-9-ylidene)amino]oxy}acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR).
J Enzyme Inhib Med Chem, 31, 2016
|
|
6DDH
 
 | | Crystal structure of the double mutant (D52N/R367Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, INOSINIC ACID | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDL
 
 | | Crystal structure of the single mutant (D52N) of NT5C2-Q523X in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
5E6Z
 
 | | Crystal structure of Ecoli Branching Enzyme with beta cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6DE2
 
 | | Crystal structure of the double mutant (D52N/L375F) of the full-length NT5C2 in the active state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
