7A78
 
 | | Crystal structure of RXR beta LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
5C65
 
 | | Structure of the human glucose transporter GLUT3 / SLC2A3 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Octyl Glucose Neopentyl Glycol, Solute carrier family 2, ... | | Authors: | Pike, A.C.W, Quigley, A, Chu, A, Tessitore, A, Xia, X, Mukhopadhyay, S, Wang, D, Kupinska, K, Strain-Damerell, C, Chalk, R, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-22 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the human glucose transporter GLUT3 / SLC2A3
To Be Published
|
|
5C8H
 
 | | Crystal structure of ORC2 C-terminal domain | | Descriptor: | Origin recognition complex subunit 2, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Dong, A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of ORC2 C-terminal domain
To Be Published
|
|
4WSQ
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, K-252A, ... | | Authors: | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Abdul, K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
6SI2
 
 | | p53 cancer mutant Y220S | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
5HTC
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | Descriptor: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6SI4
 
 | | p53 cancer mutant Y220S in complex with small-molecule stabilizer PK9323 | | Descriptor: | 1-[9-ethyl-7-(1,3-thiazol-4-yl)carbazol-3-yl]-~{N}-methyl-methanamine, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
6SI0
 
 | | p53 cancer mutant Y220C in complex with small-molecule stabilizer PK9323 | | Descriptor: | 1,2-ETHANEDIOL, 1-[9-ethyl-7-(1,3-thiazol-4-yl)carbazol-3-yl]-~{N}-methyl-methanamine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Joerger, A.C, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
5EAP
 
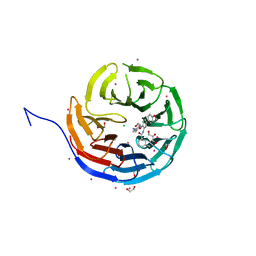 | | Crystal structure of human WDR5 in complex with compound 9e | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
6YQN
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]t rideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]benzamide | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
5EAR
 
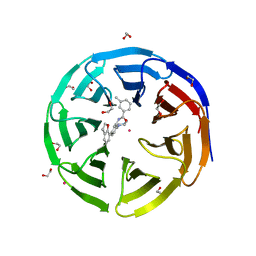 | | Crystal structure of human WDR5 in complex with compound 9d | | Descriptor: | 1,2-ETHANEDIOL, N-[5-(2,3-dihydro-1-benzofuran-7-yl)-2-(4-methylpiperazin-1-yl)phenyl]-3-methylbenzamide, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
8BZI
 
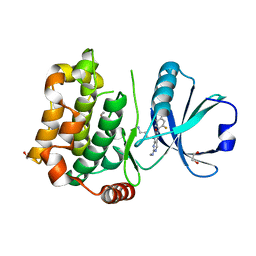 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
6SI3
 
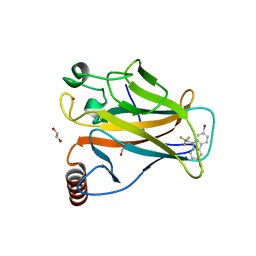 | | p53 cancer mutant Y220S in complex with small-molecule stabilizer PK9301 | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-bromanyl-9-[2,2,2-tris(fluoranyl)ethyl]carbazol-3-yl]-~{N}-methyl-methanamine, Cellular tumor antigen p53, ... | | Authors: | Joerger, A.C, Bauer, M.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
6YQO
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW12 | | Descriptor: | (S)-N1-(4-(2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetamido)phenyl)-N8-hydroxyoctanediamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
6YQP
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW22 | | Descriptor: | (~{E})-3-[4-[[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6 ),4,7,10,12-pentaen-9-yl]ethanoylamino]methyl]phenyl]-~{N}-oxidanyl-prop-2-enamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
5HTB
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3353 | | Descriptor: | (3R)-4-amino-3-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}-4-oxobutanoic acid (non-preferred name), (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-11 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5DZC
 
 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - AMPPNP bound | | Descriptor: | CHLORIDE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, He, H, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5EAM
 
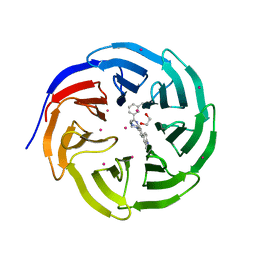 | | Crystal structure of human WDR5 in complex with compound 9o | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
6NM4
 
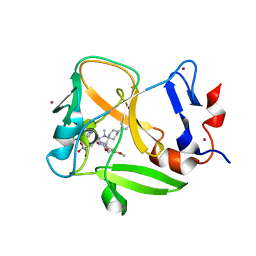 | | Crystal structure of SAM-bound PRDM9 in complex with MRK-740 inhibitor | | Descriptor: | 4-[3-(3,5-dimethoxyphenyl)-1,2,4-oxadiazol-5-yl]-1-methyl-9-(2-methylpyridin-4-yl)-1,4,9-triazaspiro[5.5]undecane, Histone-lysine N-methyltransferase PRDM9, S-ADENOSYLMETHIONINE, ... | | Authors: | Ivanochko, D, Halabelian, L, Fischer, C, Sanders, J.M, Kattar, S.D, Brown, P.J, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of a chemical probe for PRDM9.
Nat Commun, 10, 2019
|
|
8R5C
 
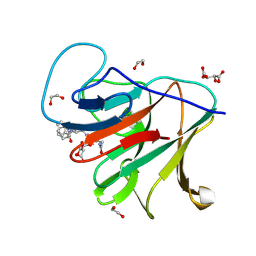 | | Crystal structure of human TRIM7 PRYSPRY domain bound to (2-(1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | (2~{S})-5-azanyl-5-oxidanylidene-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]pentanoic acid, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
5DWS
 
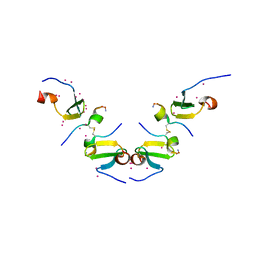 | | Crystal Structure of ITCH WW3 domain in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, UNKNOWN ATOM OR ION, txnip | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of ITCH WW3 domain in complex with TXNIP peptide
to be published
|
|
5DZD
 
 | | Crystal Structure of WW4 domain of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of WW4 domain of ITCH in complex with TXNIP peptide
To be Published
|
|
5EQ0
 
 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
6NEY
 
 | | Crystal structure of TcBDF5, a bromodomain containing protein from Trypanosoma cruzi | | Descriptor: | Uncharacterized protein | | Authors: | Loppnau, P, Dong, A, Tempel, W, Lin, Y.H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-18 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of TcBDF5, a bromodomain containing protein from Trypanosoma cruzi
To Be Published
|
|
7A77
 
 | | Crystal structure of RXR alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
