6JBJ
 
 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
6JJ5
 
 | | BRD4 in complex with 259 | | Descriptor: | 3-((1-methyl-2-oxo-1,2,2a1,5a-tetrahydro-6H-pyrido[3',2':6,7]azepino[4,3,2-cd]isoindol-6-yl)methyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | BRD4 in complex with ZZM1
To Be Published
|
|
4BV5
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 4-(aminomethyl)-N-(benzenesulfonyl)cyclohexanecarboxamide, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
1Q57
 
 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
4BVV
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
6MEY
 
 | | Crystal structure of KPC-2 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, ACETATE ION, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
5XM2
 
 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
4BVW
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1,2,3,4-tetrahydroisoquinoline-6-carboxylic acid, 1,2-ETHANEDIOL, APOLIPOPROTEIN(A), ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | Authors: | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
4DSQ
 
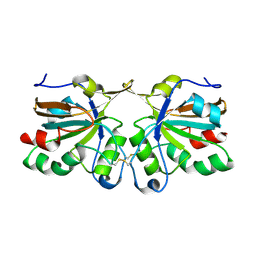 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in oxidized form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species
J.Biol.Chem., 287, 2012
|
|
6MNP
 
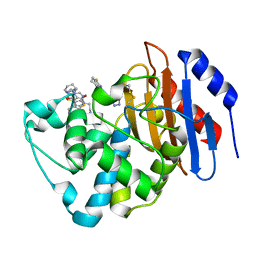 | | Crystal structure of KPC-2 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-10-02 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MLL
 
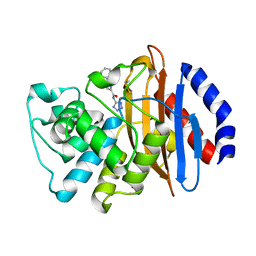 | | Crystal structure of KPC-2 with compound 7 | | Descriptor: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-27 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
4DE1
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 18 | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, N-[3-(1H-tetrazol-5-yl)phenyl]-2H-indazole-5-carboxamide | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
4DE3
 
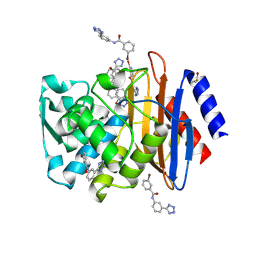 | | CTX-M-9 class A beta-lactamase complexed with compound 4 | | Descriptor: | 3-bromo-N-[3-(1H-tetrazol-5-yl)phenyl]benzamide, Beta-lactamase, DIMETHYL SULFOXIDE | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
4BBQ
 
 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
6KKN
 
 | | Crystal structure of RuBisCO accumulation factor Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
3C91
 
 | | Thermoplasma acidophilum 20S proteasome with an open gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
3EH5
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
6JY5
 
 | | Structure of CsoS4B from Halothiobacillus neapolitanus | | Descriptor: | Unidentified carboxysome polypeptide | | Authors: | Zhao, Y.Y, Jiang, Y.L, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pentameric shell protein CsoS4B of Halothiobacillus neapolitanus alpha-carboxysome.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
3XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, sorbitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
4D6V
 
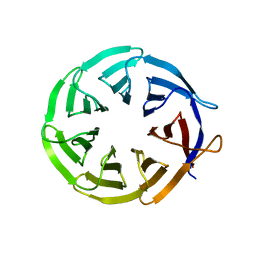 | | Crystal structure of signal transducing protein | | Descriptor: | G PROTEIN BETA SUBUNIT GIB2 | | Authors: | Ero, R, Dimitrova, V.T, Chen, Y, Bu, W, Feng, S, Liu, T, Wang, P, Xue, C, Tan, S.M, Gao, Y.G. | | Deposit date: | 2014-11-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Gib2, a Signal-Transducing Protein Scaffold Associated with Ribosomes in Cryptococcus Neoformans.
Sci.Rep., 5, 2015
|
|
3PPN
 
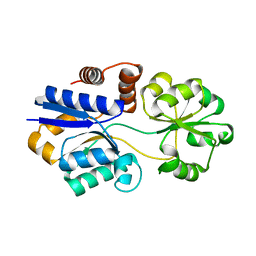 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPQ
 
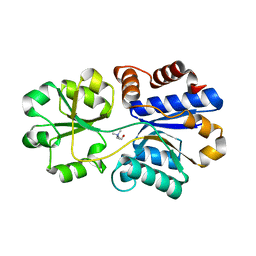 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | CHOLINE ION, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3C92
 
 | | Thermoplasma acidophilum 20S proteasome with a closed gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
2XQZ
 
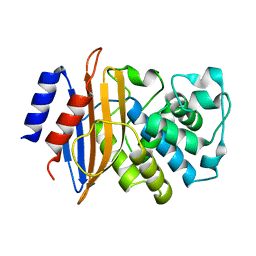 | | Neutron structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
