8IJV
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-02 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[[5-chloranyl-2-(4-chlorophenyl)phenyl]methoxy]-N-methyl-but-2-yn-1-amine, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJX
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
3DZO
 
 | | Crystal structure of a rhoptry kinase from toxoplasma gondii | | Descriptor: | MAGNESIUM ION, Rhoptry kinase domain | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Ni, S, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sibley, D, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
9FBX
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one, ... | | Authors: | Chung, C. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structure- and Property-Based Optimization of Efficient Pan-Bromodomain and Extra Terminal Inhibitors to Identify Oral and Intravenous Candidate I-BET787.
J.Med.Chem., 67, 2024
|
|
9FBY
 
 | |
8E1P
 
 | | Crystal structure of BG505 SOSIP.v4.1-GT1.2 trimer in complex with gl-PGV20 and PGT124 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505-SOSIP.v4.1-GT1.2gp120, ... | | Authors: | Sarkar, A, Kumar, S, Wilson, I.A. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.82 Å) | | Cite: | Germline-targeting HIV-1 Env vaccination induces VRC01-class antibodies with rare insertions.
Cell Rep Med, 4, 2023
|
|
6Z7G
 
 | |
6Z7F
 
 | |
3PNA
 
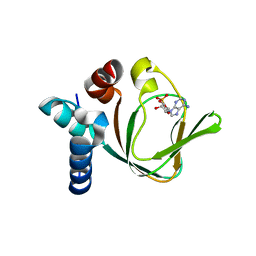 | | Crystal Structure of cAMP bound (91-244)RIa Subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Kim, C, Taylor, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Cyclic AMP Analog Blocks Kinase Activation by Stabilizing Inactive Conformation: Conformational Selection Highlights a New Concept in Allosteric Inhibitor Design.
Mol Cell Proteomics, 10, 2011
|
|
5XCC
 
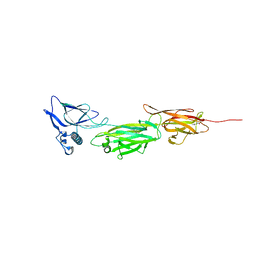 | |
5XCB
 
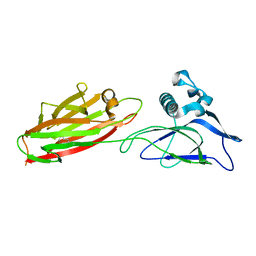 | |
6IXZ
 
 | |
6IXY
 
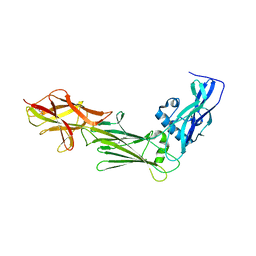 | |
7O18
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH I-BET282 | | Descriptor: | (R)-4-(8-methoxy-1-(1-methoxypropan-2-yl)-2-(tetrahydro-2H-pyran-4-yl)-1H-imidazo[4,5-c]quinolin-7-yl)-3,5-dimethylisoxazole, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Novel Bromodomain and Extra Terminal Domain (BET) Protein Inhibitor, I-BET282E, Suitable for Clinical Progression.
J.Med.Chem., 64, 2021
|
|
5U6V
 
 | | X-ray crystal structure of 1,2,3-triazolobenzodiazepine in complex with BRD2(D2) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(4-chlorophenyl)-1-methyl-6,7-dihydro-5H-[1,2,3]triazolo[1,5-d][1,4]benzodiazepin-9-yl]pyridin-2-amine, Bromodomain-containing protein 2 | | Authors: | Hatfaludi, T, Sharp, P.P, Garnier, J.-M, Burns, C.J, Czabotar, P.E. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Design, Synthesis, and Biological Activity of 1,2,3-Triazolobenzodiazepine BET Bromodomain Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
4TXW
 
 | | Crystal structure of CBM32-4 from the Clostridium perfringens NagH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-07-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution Structure and Dynamics of Full-length GH84A, a multimodular B-N-acetylglucosaminidase from Clostridium perfringens
To Be Published
|
|
2KLC
 
 | | NMR solution structure of human ubiquitin-like domain of ubiquilin 1, Northeast Structural Genomics Consortium (NESG) target HT5A | | Descriptor: | Ubiquilin-1 | | Authors: | Doherty, R.S, Dhe-Paganon, S, Fares, C, Lemak, S, Gutmanas, A, Garcia, M, Yee, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ubiquitin-like domain of ubiquilin 1
To be Published
|
|
2K7I
 
 | | Solution NMR structure of protein ATU0232 from AGROBACTERIUM TUMEFACIENS. Northeast Structural Genomics Consortium (NESG) target AtT3. Ontario Center for Structural Proteomics target ATC0223. | | Descriptor: | UPF0339 protein Atu0232 | | Authors: | Lemak, A, Srisailam, S, Yee, A, Bansal, S, Semesi, A, Prestegard, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ATU0232 from Agrobacterium Tumefaciens.
To be Published
|
|
2KVR
 
 | | Solution NMR structure of human ubiquitin specific protease Usp7 UBL domain (residues 537-664). NESG target hr4395c/ SGC-Toronto | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bezsonova, I, Lemak, A, Avvakumov, G, Xue, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | solution structure of the ubiquitin specific protease Usp7 ubiquitin-like domain
To be Published
|
|
4UYF
 
 | | N-Terminal bromodomain of Human BRD2 with I-BET726 (GSK1324726A) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2S,4R)-1-acetyl-4-[(4-chlorophenyl)amino]-2-methyl-1,2,3,4-tetrahydroquinolin-6-yl]benzoic acid, BROMODOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Gosmini, R. | | Deposit date: | 2014-08-31 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Discovery of I-Bet726 (Gsk1324726A), a Potent Tetrahydroquinoline Apoa1 Up-Regulator and Selective Bet Bromodomain Inhibitor.
J.Med.Chem., 57, 2014
|
|
4YDV
 
 | | STRUCTURE OF THE ANTIBODY 7B2 THAT CAPTURES HIV-1 VIRIONS | | Descriptor: | HIV ANTIBODY 7B2 HEAVY CHAIN,IgG H chain, HIV ANTIBODY 7B2 LIGHT CHAIN,Ig kappa chain C region, HIV GP41 PEPTIDE GP41(596-606) | | Authors: | Nicely, N.I, Pemble IV, C.W. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques.
Plos Pathog., 11, 2015
|
|
4UYH
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 1-((2R,4S)-2-methyl-4-(phenylamino)-6-(4-(piperidin-1-ylmethyl)phenyl)-3,4-dihydroquinolin-1(2H)-yl)ethanone | | Descriptor: | 1-[(2S,4R)-2-methyl-4-(phenylamino)-6-[4-(piperidin-1-ylmethyl)phenyl]-3,4-dihydroquinolin-1(2H)-yl]ethanone, BROMODOMAIN-CONTAINING PROTEIN 2, DIMETHYL SULFOXIDE, ... | | Authors: | Chung, C, Bamborough, P, Gosmini, R. | | Deposit date: | 2014-08-31 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Discovery of I-Bet726 (Gsk1324726A), a Potent Tetrahydroquinoline Apoa1 Up-Regulator and Selective Bet Bromodomain Inhibitor.
J.Med.Chem., 57, 2014
|
|
4HQQ
 
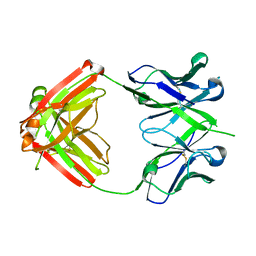 | | Crystal structure of RV144-elicited antibody CH58 | | Descriptor: | CH58 Fab heavy chain, CH58 Fab light chain | | Authors: | McLellan, J.S, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
4HPY
 
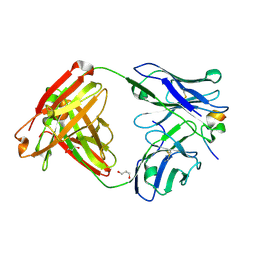 | | Crystal structure of RV144-elicited antibody CH59 in complex with V2 peptide | | Descriptor: | CH59 Fab heavy chain, CH59 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
4HPO
 
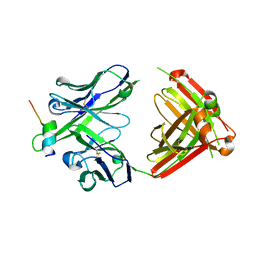 | | Crystal structure of RV144-elicited antibody CH58 in complex with V2 peptide | | Descriptor: | CH58 Fab heavy chain, CH58 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
