4MSK
 
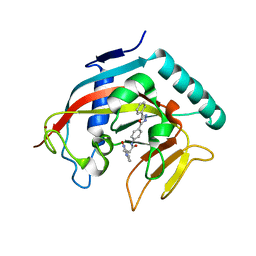 | | Co-crystal structure of tankyrase 1 with compound 34 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[4-(5-phenyl-1,3,4-oxadiazol-2-yl)phenyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MT9
 
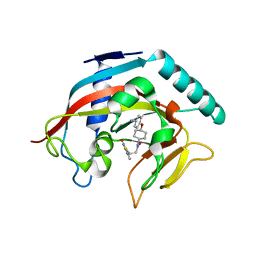 | | Co-crystal structure of tankyrase 1 with compound 49 | | Descriptor: | N-[trans-4-(4-cyanophenoxy)cyclohexyl]-3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-19 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
3BYU
 
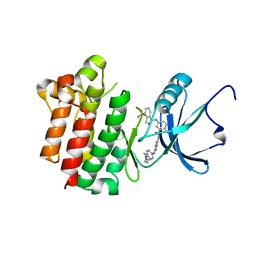 | | co-crystal structure of Lck and aminopyrimidine reverse amide 23 | | Descriptor: | 2-methyl-N-{4-methyl-3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-5-yl)carbamoyl]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of aminopyrimidine amides as potent, selective inhibitors of lymphocyte specific kinase: synthesis, structure-activity relationships, and inhibition of in vivo T cell activation.
J.Med.Chem., 51, 2008
|
|
2HK7
 
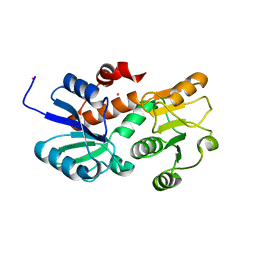 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with mercury at 2.5 angstrom resolution | | Descriptor: | MERCURY (II) ION, Shikimate dehydrogenase | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK9
 
 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with shikimate and NADP+ at 2.2 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK8
 
 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus at 2.35 angstrom resolution | | Descriptor: | Shikimate dehydrogenase | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
8Y16
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y18
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 RBD in complex with human ACE2 (local refinement from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y6A
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
6UCF
 
 | |
3BE2
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzamide inhibitor | | Descriptor: | N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Gu, Y, Rose, P, Zhao, H. | | Deposit date: | 2007-11-16 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Naphthamides as novel and potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: design, synthesis, and evaluation.
J.Med.Chem., 51, 2008
|
|
6LHK
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHA
 
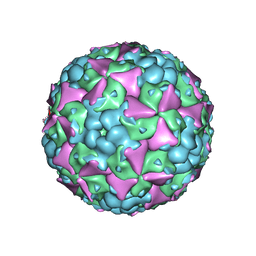 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
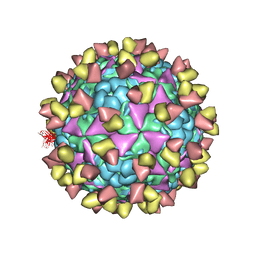 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHT
 
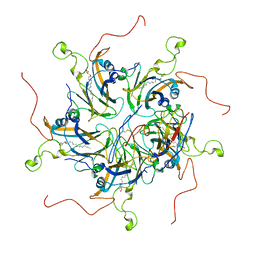 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
5QBV
 
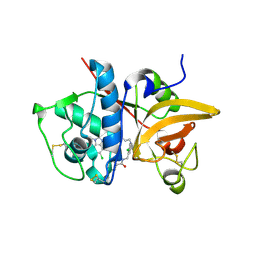 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-[2-chloro-5-(1-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-6-oxo-1,6-dihydropyridazin-3-yl)benzyl]benzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Diazinones as P2 replacements for pyrazole-based cathepsin S inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5QBY
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-[2-chloro-5-(1-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-6-oxo-1,6-dihydropyrimidin-5-yl)benzyl]-4-fluorobenzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diazinones as P2 replacements for pyrazole-based cathepsin S inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6UZ8
 
 | | Cryo-EM structure of human TRPC6 in complex with agonist AM-0883 | | Descriptor: | (5-chloro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)[(2R)-2,3-dihydro-1,4-benzodioxin-2-yl]methanone, 2-[[(2~{S})-2-decanoyloxy-3-dodecanoyloxy-propoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bai, Y, Yu, X, Huang, X, Chen, H. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for pharmacological modulation of the TRPC6 channel.
Elife, 9, 2020
|
|
6UZA
 
 | | Cryo-EM structure of human TRPC6 in complex with antagonist AM-1473 | | Descriptor: | 2-[[(2~{S})-2-decanoyloxypropoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, 4-({(1R,2R)-2-[(3R)-3-aminopiperidin-1-yl]-2,3-dihydro-1H-inden-1-yl}oxy)benzonitrile, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bai, Y, Yu, X, Huang, X, Chen, H. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for pharmacological modulation of the TRPC6 channel.
Elife, 9, 2020
|
|
6OT1
 
 | |
6OSY
 
 | |
5N8C
 
 | | Crystal structure of Pseudomonas aeruginosa LpxC complexed with inhibitor | | Descriptor: | (2~{S})-3-azanyl-2-[[(1~{R})-5-[2-[4-[[2-(hydroxymethyl)imidazol-1-yl]methyl]phenyl]ethynyl]-2,3-dihydro-1~{H}-inden-1-yl]amino]-3-methyl-~{N}-oxidanyl-butanamide, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Cross, J.B, Ryan, M.D, Zhang, J, Cheng, R.K, Wood, M, Andersen, O.A, Brooks, M, Kwong, J, Barker, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based discovery of LpxC inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2RFS
 
 | | X-ray structure of SU11274 bound to c-Met | | Descriptor: | Hepatocyte growth factor receptor, N-(3-chlorophenyl)-N-methyl-2-oxo-3-[(3,4,5-trimethyl-1H-pyrrol-2-yl)methyl]-2H-indole-5-sulfonamide | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
