2OO8
 
 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8P82
 
 | | Cryo-EM structure of dimeric UBR5 | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8P83
 
 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8PC7
 
 | |
2OFV
 
 | | crystal structure of aminoquinazoline 1 bound to Lck | | Descriptor: | 3-(2-AMINOQUINAZOLIN-6-YL)-4-METHYL-N-[3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OG8
 
 | | crystal structure of aminoquinazoline 36 bound to Lck | | Descriptor: | N-{2-[(N,N-DIETHYLGLYCYL)AMINO]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[2-(METHYLAMINO)QUINAZOLIN-6-YL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
3OKJ
 
 | | Alpha-keto-aldehyde binding mechanism reveals a novel lead structure motif for proteasome inhibition | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S,3S)-3-hydroxy-1-(4-hydroxyphenyl)-4-oxobutan-2-yl]-L-leucinamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Poynor, M, Gallastegui, P, Stein, M, Schmidt, B, Kloetzel, P.M, Huber, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elucidation of the alpha-keto-aldehyde binding mechanism: a lead structure motif for proteasome inhibition
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
7JGX
 
 | |
7JHF
 
 | |
7JGY
 
 | |
1GKA
 
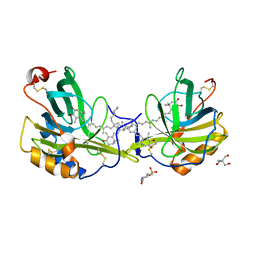 | | The molecular basis of the coloration mechanism in lobster shell. beta-crustacyanin at 3.2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASTAXANTHIN, ... | | Authors: | Cianci, M, Rizkallah, P.J, Olczak, A, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2001-08-10 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | The Molecular Basis of the Coloration Mechanism in Lobster Shell: Beta -Crustacyanin at 3.2-A Resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5HHZ
 
 | |
3FZW
 
 | | Crystal Structure of Ketosteroid Isomerase D40N-D103N from Pseudomonas putida (pKSI) with bound equilenin | | Descriptor: | EQUILENIN, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Ringe, D, Petsko, G.A. | | Deposit date: | 2009-01-26 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Hydrogen bond coupling in the ketosteroid isomerase active site.
Biochemistry, 48, 2009
|
|
6I8F
 
 | |
4R62
 
 | | Structure of Rad6~Ub | | Descriptor: | ACETATE ION, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 2 | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity.
Nucleic Acids Res., 43, 2015
|
|
5W4W
 
 | |
3DQ0
 
 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-(3-methoxy-phenyl)adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and biological activity of novel purine-derived inhibitor of cytokinin oxidase/dehydrogenase and its potential use for in vivo studies
To be Published
|
|
6RB0
 
 | |
6RKY
 
 | |
6SXP
 
 | |
1W22
 
 | | Crystal structure of inhibited human HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, N-HYDROXY-4-(METHYL{[5-(2-PYRIDINYL)-2-THIENYL]SULFONYL}AMINO)BENZAMIDE, POTASSIUM ION, ... | | Authors: | Vannini, A, Volpari, C, Caroli Casavola, E, Di Marco, S. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Eukaryotic Zn-Dependent Histone Deacetylase,Human Hdac8,Complexed with a Hydroxamic Acid Inhibitor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6SYL
 
 | |
2BQ4
 
 | | Crystal structure of type I cytochrome c3 from Desulfovibrio africanus | | Descriptor: | BASIC CYTOCHROME C3, CALCIUM ION, HEME C | | Authors: | Czjzek, M, Pieulle, L, Morelli, X, Guerlesquin, F, Hatchikian, E.C. | | Deposit date: | 2005-04-27 | | Release date: | 2005-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Type I / Type II Cytochrome C(3) Complex: An Electron Transfer Link in the Hydrogen-Sulfate Reduction Pathway.
J.Mol.Biol., 354, 2005
|
|
2V5W
 
 | | Crystal structure of HDAC8-substrate complex | | Descriptor: | GLYCYL-GLYCYL-GLYCINE, HISTONE DEACETYLASE 8, PEPTIDIC SUBSTRATE, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
3BYO
 
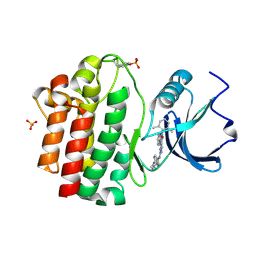 | | X-Ray co-crystal structure of 2-amino-6-phenylpyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-one 25 bound to Lck | | Descriptor: | 6-(2,6-dimethylphenyl)-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-one, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel 2-amino-6-phenyl-pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-ones as potent and orally active inhibitors of lymphocyte specific kinase (Lck): synthesis, SAR, and in vivo anti-inflammatory activity.
J.Med.Chem., 51, 2008
|
|
