9FZ6
 
 | | A 2.58A crystal structure of S. aureus DNA gyrase and DNA with metals identified through anomalous scattering | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Morgan, H, Duman, R, Bax, B.D, Warren, A.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | How Do Gepotidacin and Zoliflodacin Stabilize DNA Cleavage Complexes with Bacterial Type IIA Topoisomerases? 1. Experimental Definition of Metal Binding Sites.
Int J Mol Sci, 25, 2024
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDX
 
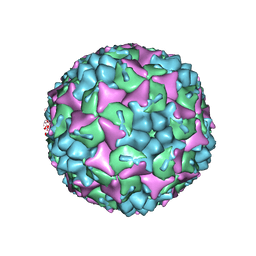 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDW
 
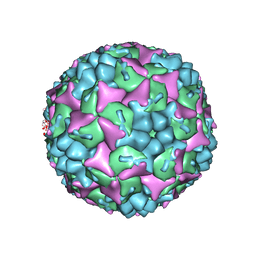 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDQ
 
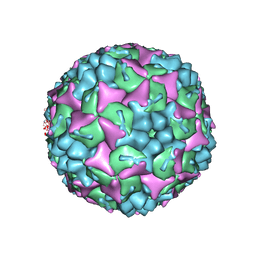 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CEW
 
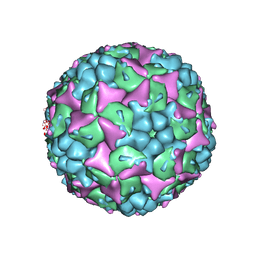 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDU
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
2XR9
 
 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
6FMF
 
 | | Neuropilin-1 b1 domain in complex with EG01377; 2.8 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1, trifluoroacetic acid | | Authors: | Yelland, T, Djordjevic, S, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
6FMC
 
 | | Neuropilin1-b1 domain in complex with EG01377, 0.9 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1 | | Authors: | Yelland, T, Djordjevic, S, Fotinou, K, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
6H3B
 
 | |
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHO
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.07 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHG
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.5 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
4G14
 
 | |
4G13
 
 | |
3QSA
 
 | | Anthranilate phosphoribosyltransferase (trpD) from Mycobacterium tuberculosis (complex with inhibitor TAMU-A7) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 4,4,4-trifluoro-1-(4-methoxyphenyl)butane-1,3-dione, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Short, F.L, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-02-20 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis anthranilate phosphoribosyltransferase by blocking of an active site entrance tunnel.
To be Published
|
|
5I4Z
 
 | | Structure of apo OmoMYC | | Descriptor: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
