8RWB
 
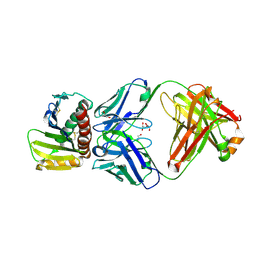 | | Crystal structure of ULBP6 in complex with a blocking antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bharill, S, Chen, I, Ivic, N, Bahrami Dizicheh, Z, Wu, Y, Doamekpor, S, Koenig, P, Fuh, G. | | Deposit date: | 2024-02-02 | | Release date: | 2025-02-12 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | 23ME-01473, an Fc Effector-Enhanced Anti-ULBP6/2/5 Antibody, Restores NK Cell-Mediated Antitumor Immunity through NKG2D and Fc gamma RIIIa Activation.
Cancer Res Commun, 5, 2025
|
|
7NB4
 
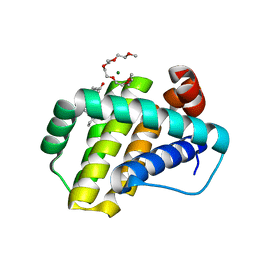 | | Structure of Mcl-1 complex with compound 1 | | Descriptor: | (2~{R})-2-[[5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
7NB7
 
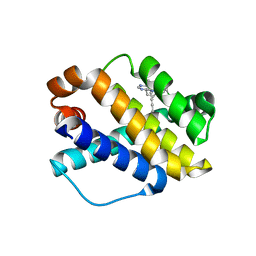 | | Structure of Mcl-1 complex with compound 6b | | Descriptor: | (2~{R})-2-[[7-but-2-ynyl-5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-pyrrolo[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
4V2Y
 
 | |
4V30
 
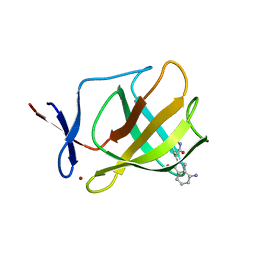 | |
4V31
 
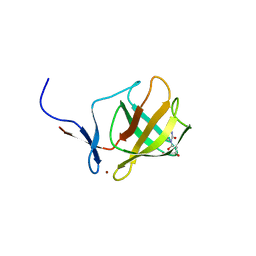 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, CEREBLON ISOFORM 4, CITRATE ANION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
4V2Z
 
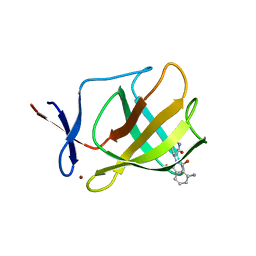 | |
9MRB
 
 | | The designed serine hydrolase known as dad_t1 | | Descriptor: | TETRAETHYLENE GLYCOL, dad_t1 | | Authors: | Pellock, S.J, Lauko, A, Bera, A, Baker, D. | | Deposit date: | 2025-01-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of serine hydrolases.
Science, 388, 2025
|
|
4V32
 
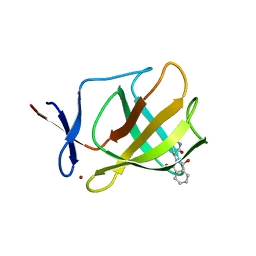 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Y101F mutant | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
3QA9
 
 | |
1YFM
 
 | |
9DEE
 
 | |
9DEH
 
 | |
9DED
 
 | | Designed protein n8 | | Descriptor: | Design protein n8 | | Authors: | Pellock, S.J, Lauko, A, Bera, A. | | Deposit date: | 2024-08-28 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Computational design of serine hydrolases.
Science, 388, 2025
|
|
9DEG
 
 | |
9DEF
 
 | |
8VC8
 
 | |
8VEI
 
 | |
8VEJ
 
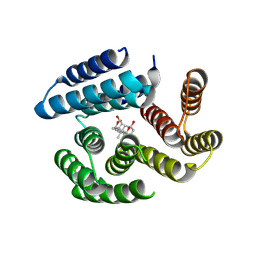 | | De novo designed cholic acid binder: CHD_buttress | | Descriptor: | CHD_buttress, CHOLIC ACID | | Authors: | Bera, A.K, Tran, L, Kang, A, Baker, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Binding and sensing diverse small molecules using shape-complementary pseudocycles.
Science, 385, 2024
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
8DT0
 
 | |
8CYK
 
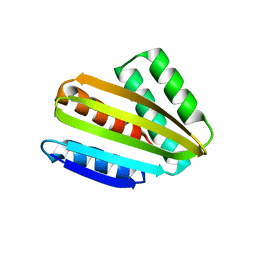 | | Crystal structure of hallucinated protein HALC1_878 | | Descriptor: | HALC1_878 | | Authors: | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
8UQF
 
 | |
5AMK
 
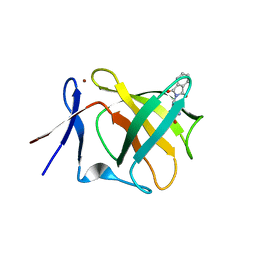 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in multiple conformations, hexagonal crystal form | | Descriptor: | CEREBLON ISOFORM 4, DIMETHYL SULFOXIDE, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5AMJ
 
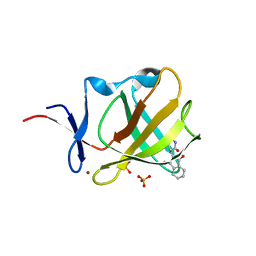 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash II structure | | Descriptor: | CEREBLON ISOFORM 4, PHOSPHATE ION, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
