5MTK
 
 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
5MMV
 
 | | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1) | | Descriptor: | (3~{S})-3-[[(3~{S})-2-[(2~{S})-3-methyl-2-(naphthalen-2-ylcarbonylamino)butanoyl]-4-oxidanyl-2-azabicyclo[2.2.2]octan-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1)
To Be Published
|
|
1MQO
 
 | |
5LWP
 
 | | Discovery of phenoxyindazoles and phenylthioindazoles as RORg inverse agonists | | Descriptor: | 4-[3-[2-chloranyl-6-(trifluoromethyl)phenoxy]-5-(dimethylcarbamoyl)indazol-1-yl]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Ouvry, G, Bouix-Peter, C, Ciesielski, F, Chantalat, L, Christin, O, Comino, C, Duvert, D, Feret, C, Harris, C.S, Luzy, A.-P, Musicki, B, Orfila, D, Pascau, J, Parnet, V, Perrin, A, Pierre, R, Raffin, C, Rival, Y, Taquet, N, Thoreau, E, Hennequin, L.F. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of phenoxyindazoles and phenylthioindazoles as ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1EEH
 
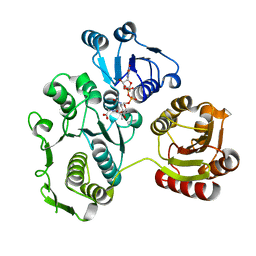 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
1E0D
 
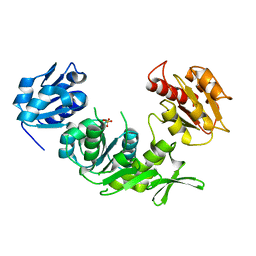 | | UDP-N-Acetylmuramoyl-L-Alanine:D-Glutamate Ligase | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Fanchon, E, Bertrand, J, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-03-24 | | Release date: | 2000-06-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Open" Structures of Murd: Domain Movements and Structural Similarities with Folylpolyglutamate Synthetase.
J.Mol.Biol., 301, 2000
|
|
5MZ6
 
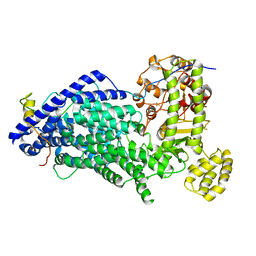 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5G04
 
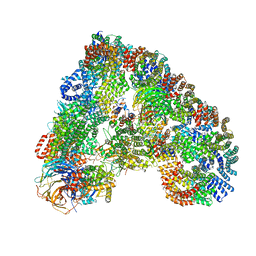 | | Structure of the human APC-Cdc20-Hsl1 complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5G05
 
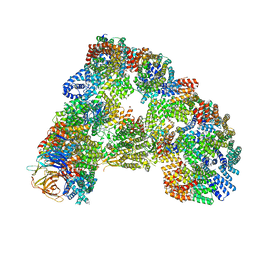 | | Cryo-EM structure of combined apo phosphorylated APC | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
7KCB
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NAD+ and Trifluoroethanol | | Descriptor: | ADH1 isoform 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-05 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KCQ
 
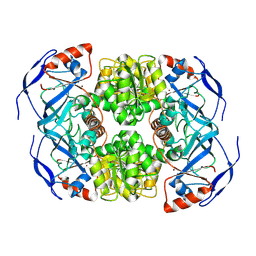 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Open Form of Apoenzyme | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KC2
 
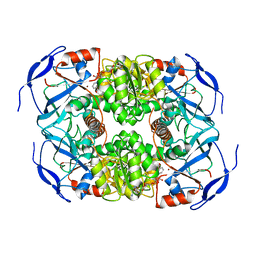 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KJY
 
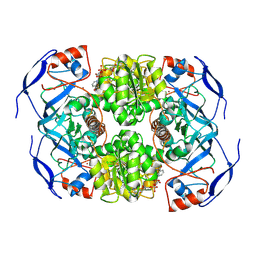 | | Symmetry in Yeast Alcohol Dehydrogenase 1 - Open Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
8S4G
 
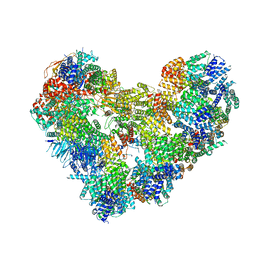 | | Cryo-EM structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2024-02-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | New structural features of the APC/C revealed by high resolution cryo-EM structures of apo-APC/C and the APC/C-CDH1-EMI1 complex
To Be Published
|
|
5LCW
 
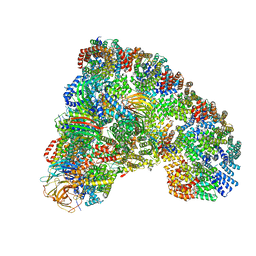 | | Cryo-EM structure of the Anaphase-promoting complex/Cyclosome, in complex with the Mitotic checkpoint complex (APC/C-MCC) at 4.2 angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Alfieri, C, Chang, L, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular basis of APC/C regulation by the spindle assembly checkpoint.
Nature, 536, 2016
|
|
1E8C
 
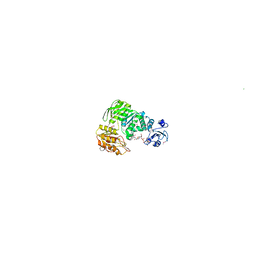 | | Structure of MurE the UDP-N-acetylmuramyl tripeptide synthetase from E. coli | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CHLORIDE ION, UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Gordon, E.J, Chantala, L, Dideberg, O. | | Deposit date: | 2000-09-19 | | Release date: | 2001-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1DYP
 
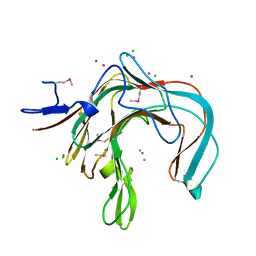 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
1S9Y
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQS, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9X
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQA, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9W
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope, SLLMWITQC, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1FFL
 
 | |
2Y6G
 
 | | Cellopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2Y6L
 
 | | Xylopentaose binding X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2Y6J
 
 | | X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2Y6H
 
 | | X-2 L110F CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
