8D7H
 
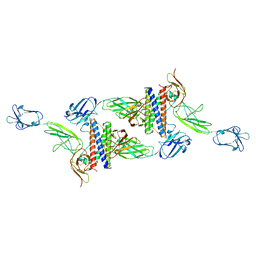 | | Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cardiotrophin-like cytokine factor 1, ... | | Authors: | Zhou, Y, Franklin, M.C. | | Deposit date: | 2022-06-07 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly of gp130 family cytokine signaling complexes.
Sci Adv, 9, 2023
|
|
8D82
 
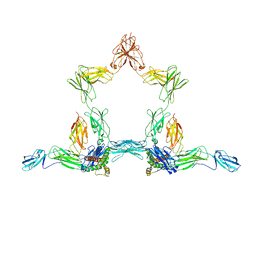 | |
8D85
 
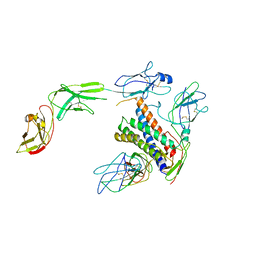 | |
5UZK
 
 | | Crystal Structure of PKA bound to an pyrrolo pyridine inhibitor | | Descriptor: | 2-{3-[3-(piperidin-4-yl)propoxy]phenyl}-N-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,3-thiazol-2-yl]acetamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Jacobs, M.D, Brown, K. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
2OJG
 
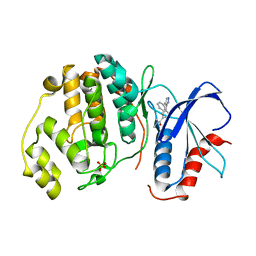 | |
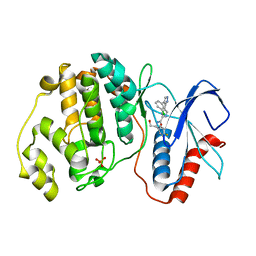
2OJJ
 
 | |
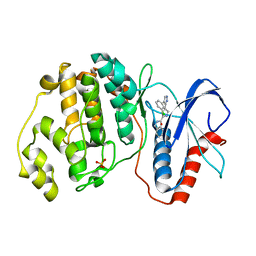
2OJI
 
 | |
2OK1
 
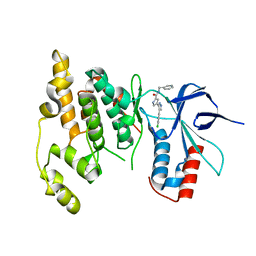 | |
5HVU
 
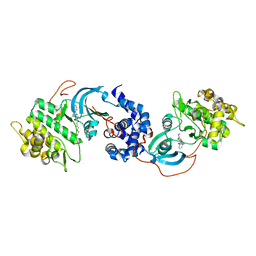 | |
7KKE
 
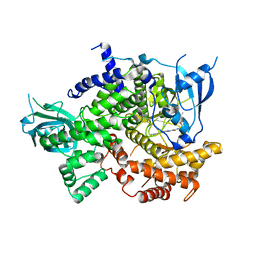 | | Phosphoinositide 3-Kinase gamma bound to a thiazole inhibitor | | Descriptor: | N-[2-(3,3-dimethylbutoxy)ethyl]-N'-{4-methyl-5-[(pyridin-4-yl)ethynyl]-1,3-thiazol-2-yl}urea, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Jacobs, M.D, Griffith, J.P. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Novel Series of Potent and Selective Alkynylthiazole-Derived PI3K gamma Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
2PST
 
 | |
8FW1
 
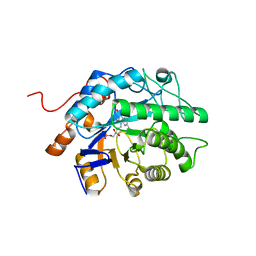 | | Gluconobacter Ene-Reductase (GluER) mutant - PagER | | Descriptor: | FLAVIN MONONUCLEOTIDE, N-ethylmaleimide reductase | | Authors: | Dahagam, S, Page, C, Patterson, M.G, Hyster, T.K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regioselective Radical Alkylation of Arenes Using Evolved Photoenzymes.
J.Am.Chem.Soc., 145, 2023
|
|
2H8N
 
 | |
7KNU
 
 | |
7KNT
 
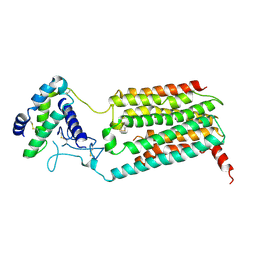 | |
3BJK
 
 | | Crystal structure of HI0827, a hexameric broad specificity acyl-coenzyme A thioesterase: The Asp44Ala mutant enzyme | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase HI0827, CITRIC ACID | | Authors: | Willis, M.A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of YciA from Haemophilus influenzae (HI0827), a Hexameric Broad Specificity Acyl-Coenzyme A Thioesterase.
Biochemistry, 47, 2008
|
|
4FA0
 
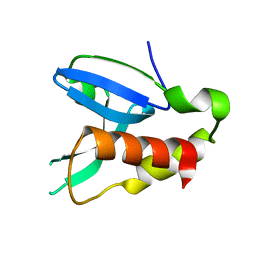 | | Crystal structure of human AdPLA to 2.65 A resolution | | Descriptor: | Group XVI phospholipase A1/A2 | | Authors: | Lovell, S, Battaile, K.P, Addington, L, Zhang, N, Rao, J.L.U.M, Moise, A.R. | | Deposit date: | 2012-05-21 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure/Function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad.
J.Biol.Chem., 287, 2012
|
|
7E4I
 
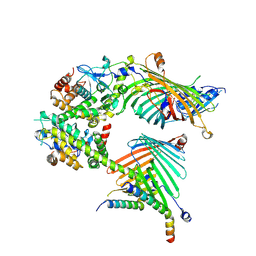 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40/Tom5/Tom6 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, Mitochondrial import receptor subunit TOM6, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7E4H
 
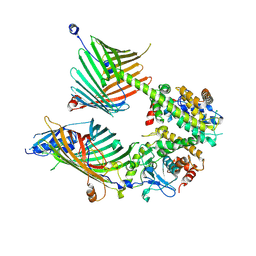 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
2O94
 
 | |
7TV3
 
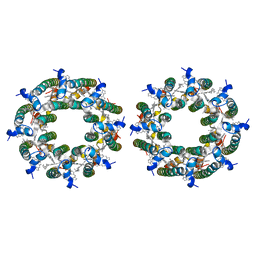 | | LH2-LH3 antenna in parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TUW
 
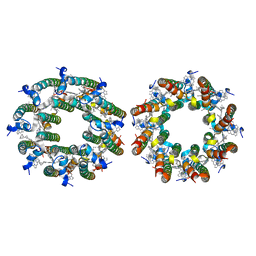 | | LH2-LH3 antenna in anti parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TNB
 
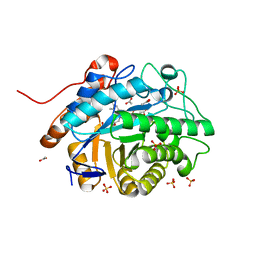 | | Caulobacter segnis arene reductase (CSAR) - WT | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase, ... | | Authors: | Garfinkle, S.E, Jeffrey, P, Hyster, T.K. | | Deposit date: | 2022-01-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An asymmetric sp 3 -sp 3 cross-electrophile coupling using 'ene'-reductases.
Nature, 610, 2022
|
|
6C1S
 
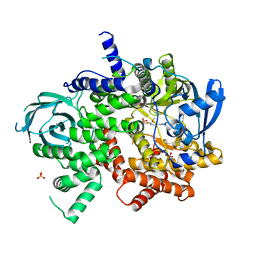 | | Phosphoinositide 3-Kinase gamma bound to an pyrrolopyridinone Inhibitor | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, {4-[2-(5,6-dimethoxypyridin-3-yl)-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-1H-pyrazol-1-yl}acetonitrile | | Authors: | Jacobs, M.D, Griffin, J.P. | | Deposit date: | 2018-01-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Synthesis of a Novel Series of Orally Bioavailable, CNS-Penetrant, Isoform Selective Phosphoinositide 3-Kinase gamma (PI3K gamma ) Inhibitors with Potential for the Treatment of Multiple Sclerosis (MS).
J. Med. Chem., 61, 2018
|
|
8FB9
 
 | | LH2-LH3 antenna in anti parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-11-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
