6U28
 
 | | Crystal structure of 1918 NS1-ED W187A in complex with the p85-beta-iSH2 domain of human PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Cho, J.H, Zhao, B, Savage, N, Li, P. | | Deposit date: | 2019-08-19 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8I8Y
 
 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4JQ6
 
 | | Crystal structure of blue light-absorbing proteorhodopsin from Med12 at 2.3 Angstrom | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Proteorhodopsin, RETINAL | | Authors: | Ozorowski, G, Luecke, H. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6L4R
 
 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
2YEZ
 
 | | COMPLEX OF A B21 CHICKEN MHC CLASS I MOLECULE AND A 10MER CHICKEN PEPTIDE | | Descriptor: | 14-3-3 PROTEIN THETA, BETA-2-MICROGLOBULIN, MAJOR HISTOCOMPATIBILITY COMPLEX CLASS I GLYCOPROTEIN HAPLOTYPE B21 | | Authors: | Chappell, P, Roversi, P, Harrison, M.C, Mears, L.E, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.
Elife, 4, 2015
|
|
8IM6
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, Y.R, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
4I9S
 
 | |
4I9R
 
 | |
8JF4
 
 | | The crystal structure of human AURKA kinase domain in complex with AURKA-compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Aurora kinase A | | Authors: | Zhu, C.J. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89288354 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JF3
 
 | | C-Src in complex with compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Zhang, Z.M, Huang, H.S. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.84647632 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7E36
 
 | | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkanesulfonate monooxygenase SsuD/methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (Luciferase family), ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis.
Nat Commun, 12, 2021
|
|
7EW7
 
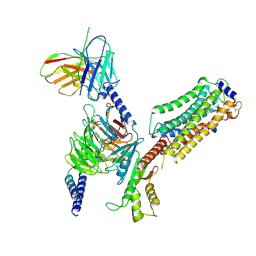 | | Cryo-EM structure of SEW2871-bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 5-[4-phenyl-5-(trifluoromethyl)thiophen-2-yl]-3-[3-(trifluoromethyl)phenyl]-1,2,4-oxadiazole, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EW1
 
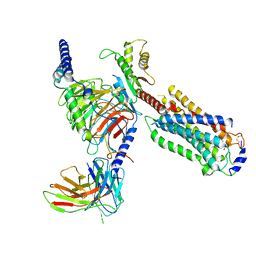 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 5 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EW0
 
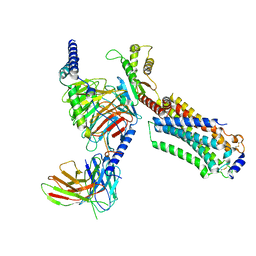 | | Cryo-EM structure of ozanimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 5-[3-[(1~{S})-1-(2-hydroxyethylamino)-2,3-dihydro-1~{H}-inden-4-yl]-1,2,4-oxadiazol-5-yl]-2-propan-2-yloxy-benzenecarbonitrile, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EVZ
 
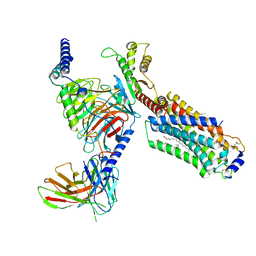 | | Cryo-EM structure of cenerimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | (2~{S})-3-[4-[5-(2-cyclopentyl-6-methoxy-pyridin-4-yl)-1,2,4-oxadiazol-3-yl]-2-ethyl-6-methyl-phenoxy]propane-1,2-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EVY
 
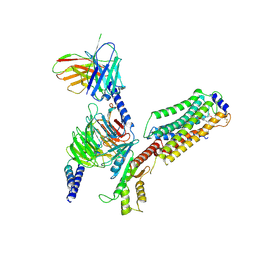 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
6KAF
 
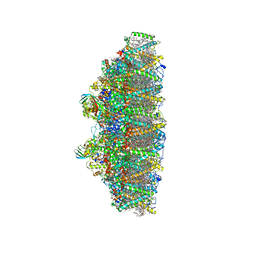 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7EFR
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain in complex with high affinity ACE2 mutant (T27W,N330Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Lu, G.W, Ye, F, Lin, X. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | S19W, T27W, and N330Y mutations in ACE2 enhance SARS-CoV-2 S-RBD binding toward both wild-type and antibody-resistant viruses and its molecular basis.
Signal Transduct Target Ther, 6, 2021
|
|
7EFP
 
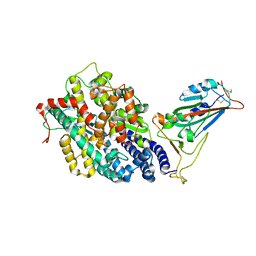 | | Structure of SARS-CoV-2 spike receptor-binding domain in complex with high affinity ACE2 mutant (S19W,N330Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Lu, G.W, Ye, F, Lin, X. | | Deposit date: | 2021-03-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | S19W, T27W, and N330Y mutations in ACE2 enhance SARS-CoV-2 S-RBD binding toward both wild-type and antibody-resistant viruses and its molecular basis.
Signal Transduct Target Ther, 6, 2021
|
|
4KZJ
 
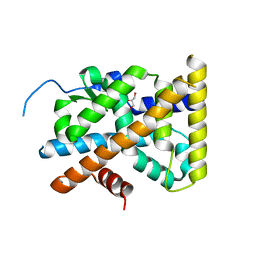 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZI
 
 | | Crystal Structure of TR3 LBD in complex with DPDO | | Descriptor: | 1-(3,5-dimethoxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
5XV7
 
 | | SRPK1 in complex with Alectinib | | Descriptor: | 1,2-ETHANEDIOL, 9-ethyl-6,6-dimethyl-8-[4-(morpholin-4-yl)piperidin-1-yl]-11-oxo-6,11-dihydro-5H-benzo[b]carbazole-3-carbonitrile, serine-arginine (SR) protein kinase 1 | | Authors: | Zeng, C, Ngo, J.C.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | SRPKIN-1: A Covalent SRPK1/2 Inhibitor that Potently Converts VEGF from Pro-angiogenic to Anti-angiogenic Isoform
Cell Chem Biol, 25, 2018
|
|
7BOK
 
 | | Cryo-EM structure of the encapsulated DyP-type peroxidase from Mycobacterium smegmatis | | Descriptor: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BOJ
 
 | | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | | Descriptor: | 29 kDa antigen Cfp29 | | Authors: | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
