1JB8
 
 | | The Crystal Structure of an RNA/DNA Hybrid Reveals Novel Intermolecular Intercalation | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3' | | Authors: | Han, G.W, Kopka, M.L, Langs, D, Dickerson, R.E. | | Deposit date: | 2001-06-02 | | Release date: | 2003-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of an RNADNA hybrid reveals intermolecular
intercalation: Dimer formation by base-pair swapping
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1JBH
 
 | | Solution structure of cellular retinol binding protein type-I in the ligand-free state | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-06-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of Apo- and holo-cellular retinol-binding protein in solution.
J.Biol.Chem., 277, 2002
|
|
5AWT
 
 | |
1JAA
 
 | | Solution structure of lactam analogue (DapE) of HIV gp41 600-612 loop. | | Descriptor: | DapE : (Ace)IWG(Dap)SGKLIETTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-30 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
6YEY
 
 | | Xenorhabdus nematophila XptA1 in complex with porcine mucosa heparin | | Descriptor: | A component of insecticidal toxin complex (Tc) | | Authors: | Roderer, D, Broecker, F, Sitsel, O, Kaplonek, P, Leidreiter, F, Seeberger, P.H, Raunser, S. | | Deposit date: | 2020-03-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Glycan-dependent cell adhesion mechanism of Tc toxins.
Nat Commun, 11, 2020
|
|
5AZ9
 
 | |
5BPT
 
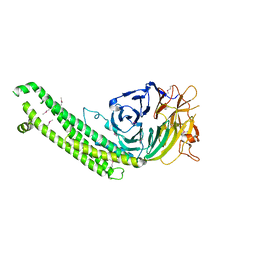 | |
1JDH
 
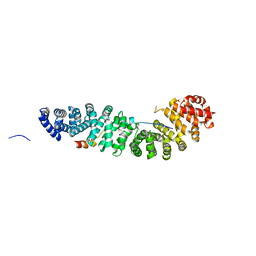 | | CRYSTAL STRUCTURE OF BETA-CATENIN AND HTCF-4 | | Descriptor: | BETA-CATENIN, hTcf-4 | | Authors: | Graham, T.A, Ferkey, D.M, Mao, F, Kimelman, D, Xu, W. | | Deposit date: | 2001-06-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tcf4 can specifically recognize beta-catenin using alternative conformations.
Nat.Struct.Biol., 8, 2001
|
|
5AO1
 
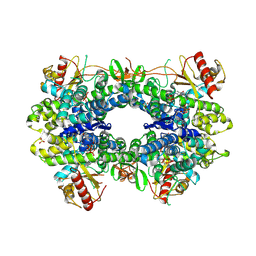 | |
5AQV
 
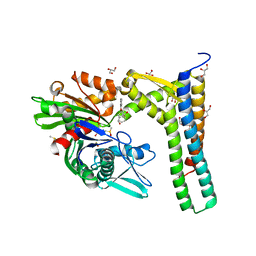 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (1R,2S,3R,5R)-3-((5-(benzyloxy)quinazolin-4-yl)amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQJ
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9H-purine-6,8-diamine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQF
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQS
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, GLYCEROL, HEAT SHOCK COGNATE 71 KDA PROTEIN, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
4M5T
 
 | | Disulfide trapped human alphaB crystallin core domain in complex with C-terminal peptide | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Cascio, D, Hochberg, G, Sawaya, M.R, Benesch, J.L.P, Robinson, C.V, Eisenberg, D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1JB2
 
 | | CRYSTAL STRUCTURE OF NTF2 M84E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
6GQE
 
 | |
5AQP
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
1JBM
 
 | |
6YNK
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_068 | | Descriptor: | 6-[[furan-2-ylmethyl(methyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNN
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_135 | | Descriptor: | 6-[[(2-chloranyl-6-fluoranyl-phenyl)methyl-methyl-amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
5BUW
 
 | |
1JC8
 
 | | Solution structure of lactam analogue (DDap) of gp41 600-612 loop of HIV | | Descriptor: | DDap: (ACE)IWGDSGKLI(DNP)TTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-08 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1JCP
 
 | | Solution structure of the lactam analogue EDap of HIV gp41 600-612 loop. | | Descriptor: | Edap : ACE-Ile-Trp-Glu-Ser-Gly-Lys-Leu-Ile-Dap-Thr-Thr-Ala ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-11 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
6YLR
 
 | | Translation initiation factor 4E in complex with bn7GpppG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, bn7GpppG mRNA 5' cap analog | | Authors: | Kubacka, D, Wojcik, R, Baranowski, M.R, Warminski, M, Kowalska, J, Jemielity, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1954546 Å) | | Cite: | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6YM0
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | Descriptor: | Spike glycoprotein, heavy chain, light chain | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
