8WO3
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Mupirocin | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNF
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in apo form | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNI
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WO2
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNJ
 
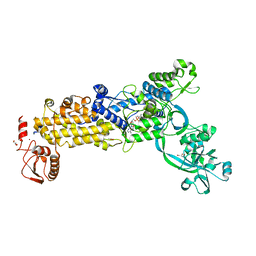 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8Y31
 
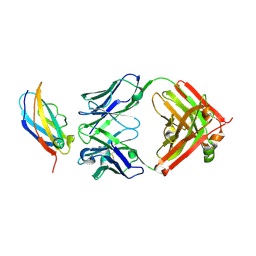 | | The crystal structure of the QX006N-Fab/IFNAR1-SD123 complex | | Descriptor: | Interferon alpha/beta receptor 1, QX006N-Fab-HC, QX006N-Fab-LC | | Authors: | Li, W, Feng, W. | | Deposit date: | 2024-01-28 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the recognition of IFNAR1 by the humanized therapeutic monoclonal antibody QX006N for the treatment of systemic lupus erythematosus.
Int.J.Biol.Macromol., 268, 2024
|
|
8Y2K
 
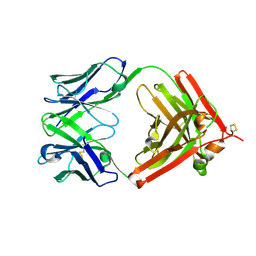 | | The crystal structure of QX006N-Fab | | Descriptor: | QX006N-Fab-HC, QX006N-Fab-LC | | Authors: | Li, W, Feng, W. | | Deposit date: | 2024-01-26 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural basis for the recognition of IFNAR1 by the humanized therapeutic monoclonal antibody QX006N for the treatment of systemic lupus erythematosus.
Int.J.Biol.Macromol., 268, 2024
|
|
7V29
 
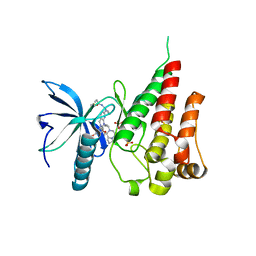 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-08-07 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
7E6T
 
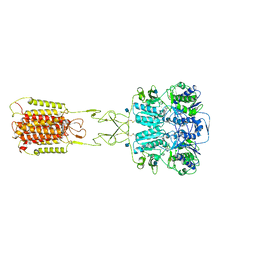 | | Structural insights into the activation of human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYCLOMETHYLTRYPTOPHAN, ... | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7E6U
 
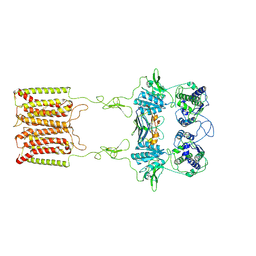 | | the complex of inactive CaSR and NB2D11 | | Descriptor: | Extracellular calcium-sensing receptor, NB-2D11 | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
6IHG
 
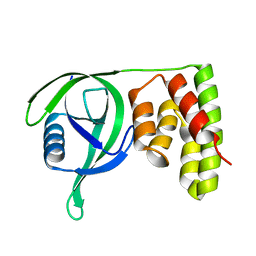 | | N terminal domain of Mycobacterium avium complex Lon protease | | Descriptor: | Lon protease | | Authors: | Chen, X.Y, Zhang, S.J, Bi, F.K, Guo, C.Y, Yao, H.W, Lin, D.H. | | Deposit date: | 2018-09-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
8IDH
 
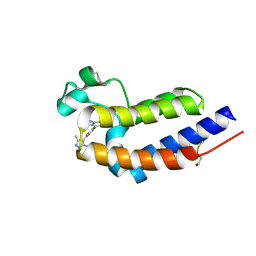 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
7XXJ
 
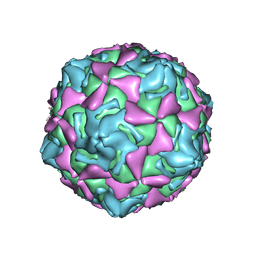 | | Echo 18 incubated with FcRn at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7XXA
 
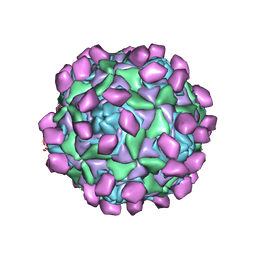 | | Complex of Echo 18 and FcRn at pH7.4 | | Descriptor: | IgG receptor FcRn large subunit p51, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7XXG
 
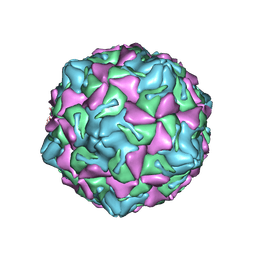 | | Echo 18 at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7EMF
 
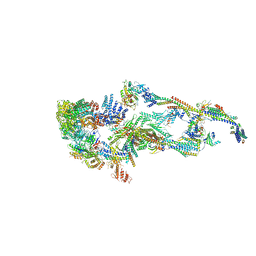 | | Human Mediator (deletion of MED1-IDR) in a Tail-extended conformation | | Descriptor: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 16, Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | Deposit date: | 2021-04-13 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENJ
 
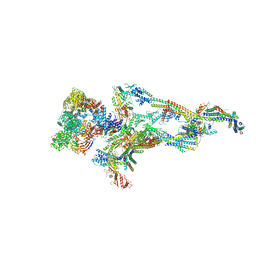 | | Human Mediator (deletion of MED1-IDR) in a Tail-bent conformation (MED-B) | | Descriptor: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, ... | | Authors: | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | Deposit date: | 2021-04-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7FJM
 
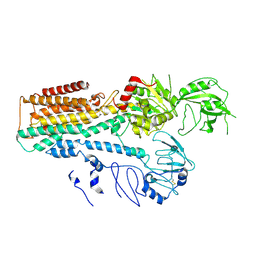 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | Polyamine-transporting ATPase 13A2 | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7F65
 
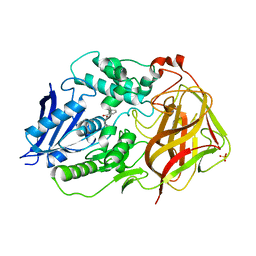 | | Bacetrial Cocaine Esterase with mutations T172R/G173Q/V116K/S117A/A51L, bound to benzoic acid | | Descriptor: | BENZOIC ACID, Cocaine esterase, SULFATE ION | | Authors: | Ouyang, P.F, Zhang, Y, Tong, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Computational Design and Crystal Structure of a Highly Efficient Benzoylecgonine Hydrolase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5IT2
 
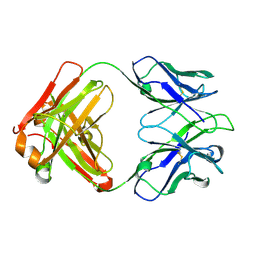 | | Structure of a transglutaminase 2-specific autoantibody 693-10-B06 Fab fragment | | Descriptor: | heavy chain, light chain | | Authors: | Chen, X, Dalhus, B, Hnida, K, Iversen, R, Sollid, L.M. | | Deposit date: | 2016-03-16 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions
Nat. Med., 18, 2012
|
|
1KVX
 
 | |
2I4W
 
 | | HIV-1 protease WT with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4D
 
 | | Crystal structure of WT HIV-1 protease with GS-8373 | | Descriptor: | ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONIC ACID, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-21 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4V
 
 | | HIV-1 protease I84V, L90M with TMC126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4U
 
 | | HIV-1 protease with TMC-126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
