3QET
 
 | |
3QEV
 
 | |
8PYI
 
 | | Human IGF1R with inhibitor 6 | | Descriptor: | 3-[8-azanyl-1-(4-ethoxy-8-fluoranyl-2-phenyl-quinolin-7-yl)imidazo[1,5-a]pyrazin-3-yl]-1-methyl-cyclobutan-1-ol, Insulin-like growth factor 1 receptor beta chain | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Human IGF1R with inhibitor 6
To Be Published
|
|
8PYJ
 
 | | Human IGF1R with inhibitor 8 | | Descriptor: | 5,5-dimethyl-1-(quinolin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Human IGF1R with inhibitor 8
To Be Published
|
|
8PYK
 
 | | Human IGF1R with inhibitor 47 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, Insulin-like growth factor 1 receptor beta chain, NICKEL (II) ION | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human IGF1R with inhibitor 47
To Be Published
|
|
8PYM
 
 | | Human IGF1R with inhibitor 54 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Human IGF1R with inhibitor 54
To Be Published
|
|
8PYN
 
 | | Human IGF1R with inhibitor 56 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Human IGF1R with inhibitor 56
To Be Published
|
|
8PYL
 
 | | Human IGF1R with inhibitor 53 | | Descriptor: | (5S)-5-methyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Human IGF1R with inhibitor 53
To Be Published
|
|
7D72
 
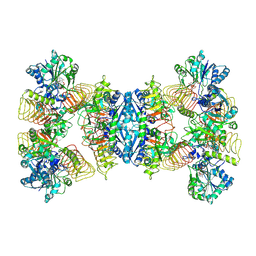 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D73
 
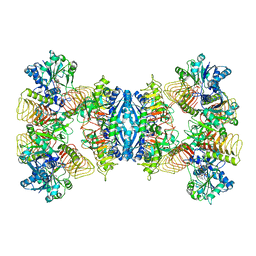 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D74
 
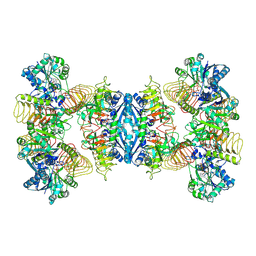 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (state II) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, Mannose-1-phosphate guanyltransferase beta | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1NJ2
 
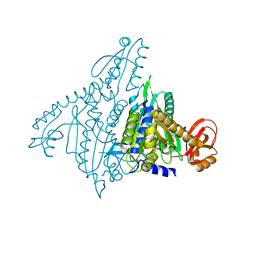 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus | | Descriptor: | MAGNESIUM ION, Proline-tRNA Synthetase, ZINC ION | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6A7H
 
 | | Bacterial protein toxins | | Descriptor: | RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
5NDI
 
 | |
5NEP
 
 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
6C6F
 
 | |
6A8J
 
 | | Crystal structure of bacterial protein toxins | | Descriptor: | GLYCEROL, RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
3JVG
 
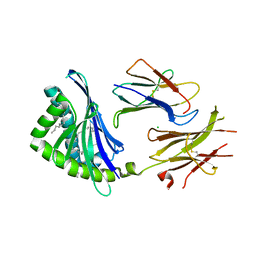 | | Crystal Structure of chicken CD1-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Dvir, H, Wang, J, Zajonc, D.M. | | Deposit date: | 2009-09-16 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for lipid-antigen recognition in avian immunity.
J.Immunol., 184, 2010
|
|
3P49
 
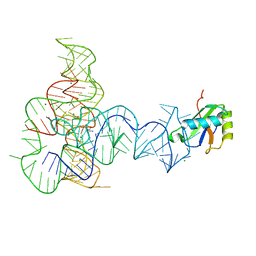 | | Crystal Structure of a Glycine Riboswitch from Fusobacterium nucleatum | | Descriptor: | GLYCINE, GLYCINE RIBOSWITCH, MAGNESIUM ION, ... | | Authors: | Butler, E.B, Wang, J, Xiong, Y, Strobel, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of cooperative ligand binding by the glycine riboswitch.
Chem.Biol., 18, 2011
|
|
5O62
 
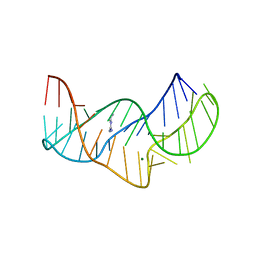 | |
5O69
 
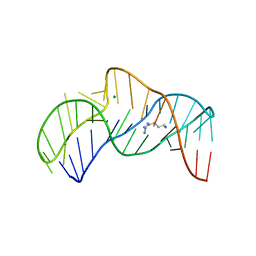 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
1NGM
 
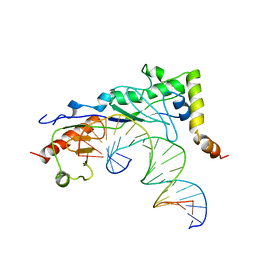 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
2F3V
 
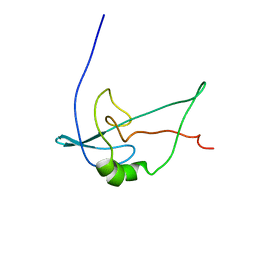 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
5X6G
 
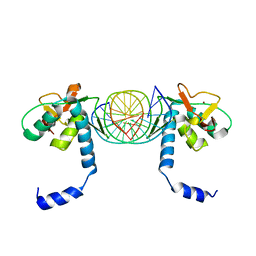 | | Crystal Structure of SMAD5-MH1/palindromic SBE DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
5X6M
 
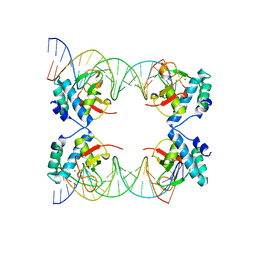 | | Crystal Structure of SMAD5-MH1 in complex with a composite DNA sequence | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
