3GP5
 
 | |
2V58
 
 | | CRYSTAL STRUCTURE OF BIOTIN CARBOXYLASE FROM E.COLI IN COMPLEX WITH POTENT INHIBITOR 1 | | Descriptor: | 6-(2,6-dibromophenyl)pyrido[2,3-d]pyrimidine-2,7-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Class of Selective Antibacterials Derived from a Protein Kinase Inhibitor Pharmacophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4Z76
 
 | |
3FLU
 
 | |
4ZJV
 
 | |
2XTK
 
 | |
7Z3Z
 
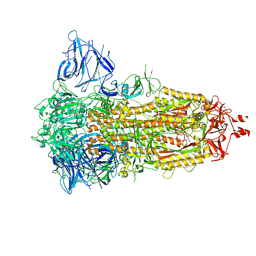 | | Locked Wuhan SARS-CoV2 Prefusion Spike ectodomain with lipid bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, STEARIC ACID, ... | | Authors: | Duyvesteyn, H.M.E, Carrique, L, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2022-03-03 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer
bioRxiv, 2020
|
|
2WD8
 
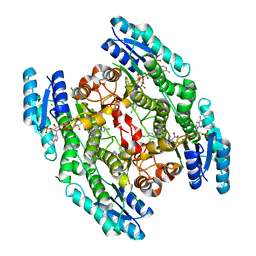 | | PTERIDINE REDUCTASE 1 (PTR1) FROM TRYPANOSOMA BRUCEI IN COMPLEX WITH NADP AND DDD00071204 | | Descriptor: | 1-(3,4-DICHLOROBENZYL)-7-PHENYL-1H-BENZIMIDAZOL-2-AMINE, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Robinson, D.A, Wyatt, P.G, Spinks, D, Brenk, R. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | One Scaffold, Three Binding Modes: Novel and Selective Pteridine Reductase 1 Inhibitors Derived from Fragment Hits Discovered by Virtual Screening.
J.Med.Chem., 52, 2009
|
|
2VRP
 
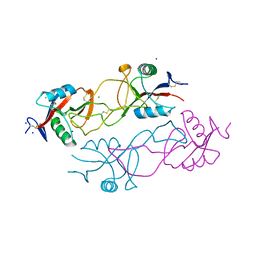 | | Structure of rhodocytin | | Descriptor: | AGGRETIN ALPHA CHAIN, AGGRETIN BETA CHAIN, CHLORIDE ION, ... | | Authors: | Watson, A.A, O'Callaghan, C.A. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of Rhodocytin, a Ligand for the Platelet-Activating Receptor Clec-2.
Protein Sci., 17, 2008
|
|
3GXN
 
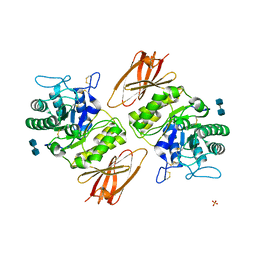 | | Crystal structure of apo alpha-galactosidase A at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, SULFATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
2WD7
 
 | | PTERIDINE REDUCTASE 1 (PTR1) FROM TRYPANOSOMA BRUCEI IN COMPLEX WITH NADP AND DDD00066750 | | Descriptor: | 6-chloro-1H-benzimidazol-2-amine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | Authors: | Robinson, D.A, Wyatt, P.G, Spinks, D, Brenk, R. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | One Scaffold, Three Binding Modes: Novel and Selective Pteridine Reductase 1 Inhibitors Derived from Fragment Hits Discovered by Virtual Screening.
J.Med.Chem., 52, 2009
|
|
3GXI
 
 | | Crystal structure of acid-beta-glucosidase at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3H0J
 
 | |
7K7Q
 
 | |
7K7O
 
 | |
3GOM
 
 | | Barium bound to the Holliday junction sequence d(TCGGCGCCGA)4 | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3', BARIUM ION | | Authors: | Naseer, A, Cardin, C.J. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3GOD
 
 | |
3GPR
 
 | | Crystal structure of rhodocetin | | Descriptor: | Rhodocetin subunit alpha, Rhodocetin subunit beta, Rhodocetin subunit delta, ... | | Authors: | Stetefeld, J. | | Deposit date: | 2009-03-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The alpha2beta1 integrin-specific antagonist rhodocetin is a cruciform, heterotetrameric molecule
Faseb J., 23, 2009
|
|
7K77
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025
To Be Published
|
|
3GXP
 
 | | Crystal structure of acid-alpha-galactosidase A complexed with galactose at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3H3C
 
 | | Crystal structure of PYK2 in complex with Sulfoximine-substituted trifluoromethylpyrimidine analog | | Descriptor: | 4-{[4-{[(1R,2R)-2-(dimethylamino)cyclopentyl]amino}-5-(trifluoromethyl)pyrimidin-2-yl]amino}-N-methylbenzenesulfonamide, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S, Mistry, A. | | Deposit date: | 2009-04-16 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfoximine-substituted trifluoromethylpyrimidine analogs as inhibitors of proline-rich tyrosine kinase 2 (PYK2) show reduced hERG activity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GXF
 
 | |
3H0S
 
 | |
3GXT
 
 | |
2YN8
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE | | Authors: | Read, J, Brassington, C.A, Overmann, R. | | Deposit date: | 2012-10-13 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Stability and Solubility Engineering of the Ephb4 Tyrosine Kinase Catalytic Domain Using a Rationally Designed Synthetic Library.
Protein Eng.Des.Sel., 26, 2013
|
|
