5YST
 
 | | RanM189D in complex with RanBP1-CRM1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1,Exportin-1, ... | | Authors: | Sun, Q, Zhang, Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | RanM189D in complex with RanBP1-CRM1
To Be Published
|
|
5YSU
 
 | | Plumbagin in complex with CRM1-RanM189D-RanBP1 | | Descriptor: | (2~{R})-2-methyl-5-oxidanyl-2,3-dihydronaphthalene-1,4-dione, CHLORIDE ION, Exportin-1,Exportin-1, ... | | Authors: | Sun, Q, Zhang, Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RanL182A in complex with RanBP1-CRM1
To Be Published
|
|
5YIX
 
 | | Caulobacter crescentus GcrA sigma-interacting domain (SID) in complex with domain 2 of sigma 70 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, RNA polymerase sigma factor RpoD, ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YRO
 
 | | RanL182A in complex with RanBP1-CRM1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1,Exportin-1, ... | | Authors: | Sun, Q, Zhang, Y. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | RanL182A in complex with RanBP1-CRM1
To Be Published
|
|
5YTB
 
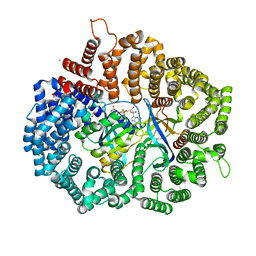 | | RanY197A in complex with RanBP1-CRM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Zhang, Y. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RanY197A in complex with RanBP1-CRM1
To Be Published
|
|
8JCH
 
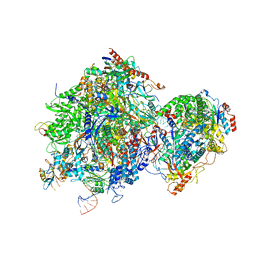 | |
2LD7
 
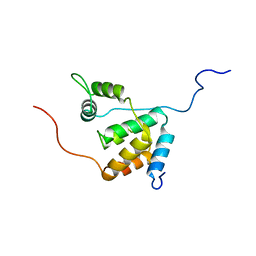 | | Solution structure of the mSin3A PAH3-SAP30 SID complex | | Descriptor: | Histone deacetylase complex subunit SAP30, Paired amphipathic helix protein Sin3a | | Authors: | Xie, T, He, Y, Korkeamaki, H, Zhang, Y, Imhoff, R, Lohi, O, Radhakrishnan, I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding.
J.Biol.Chem., 286, 2011
|
|
5ZX2
 
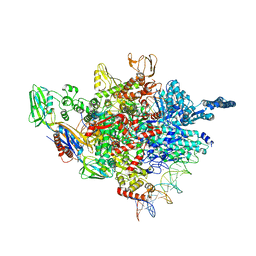 | |
8K5P
 
 | |
8KCB
 
 | |
8KCC
 
 | |
6V4P
 
 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | Descriptor: | Abciximab, heavy chain, light chain, ... | | Authors: | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
3TDB
 
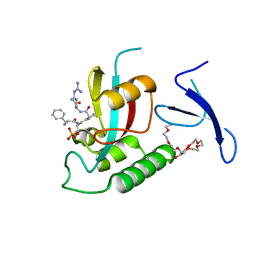 | | Human Pin1 bound to trans peptidomimetic inhibitor | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, N-[(1E,2R)-1-[(2R)-2-{[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]carbamoyl}cyclopentylidene]-3-(phosphonooxy)propan-2-yl]-L-phenylalaninamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2011-08-10 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Structural and Kinetic Analysis of Prolyl-isomerization/Phosphorylation Cross-Talk in the CTD Code.
Acs Chem.Biol., 7, 2012
|
|
6L4R
 
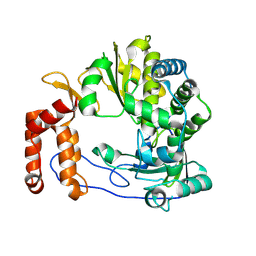 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
6VG7
 
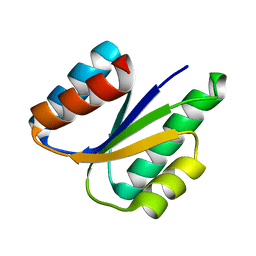 | |
2M19
 
 | | Solution structure of the Haloferax volcanii HVO 2177 protein | | Descriptor: | Molybdopterin converting factor subunit 1 | | Authors: | Li, Y, Maciejewski, M.W, Martin, J, Jin, K, Zhang, Y, Lu, M, Maupin-Furlow, J.A, Hao, B. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7JVQ
 
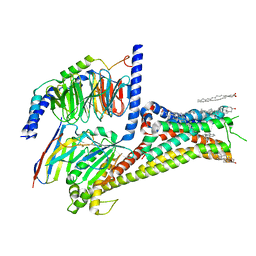 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVP
 
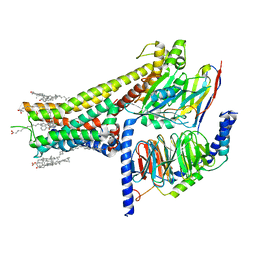 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
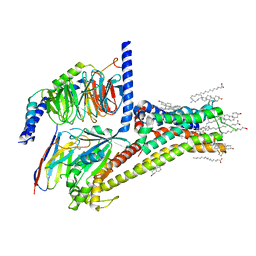 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVR
 
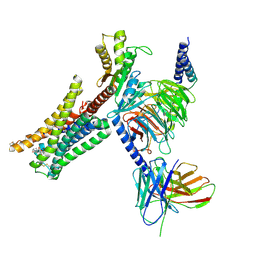 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
8HNC
 
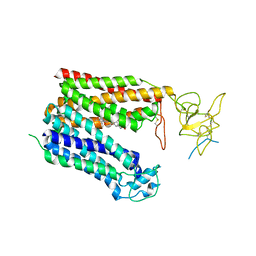 | | Cryo-EM structure of human OATP1B1 in complex with bilirubin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HND
 
 | | Cryo-EM structure of human OATP1B1 in complex with estrone-3-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
3TCZ
 
 | | Human Pin1 bound to cis peptidomimetic inhibitor | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, N~2~-({(1R,2Z)-2-[(2R)-2-(formylamino)-3-(phosphonooxy)propylidene]cyclopentyl}carbonyl)-L-argininamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Kinetic Analysis of Prolyl-isomerization/Phosphorylation Cross-Talk in the CTD Code.
Acs Chem.Biol., 7, 2012
|
|
6BQ1
 
 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
