5Y2Y
 
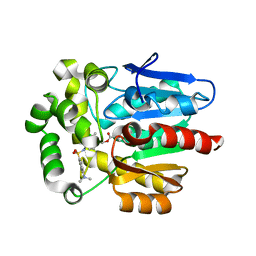 | | Crystal structure of HaloTag (M175C) complexed with dansyl-PEG2-HaloTag ligand | | Descriptor: | 5-(dimethylamino)-~{N}-[2-(2-hexoxyethoxy)ethyl]naphthalene-1-sulfonamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
6WKZ
 
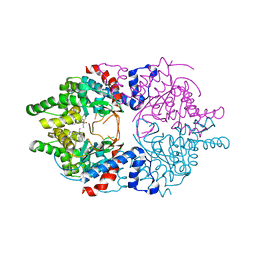 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(1R)-2-AMINO-1-PHENYLETHYL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(1R)-2-amino-1-phenylethyl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
6VEH
 
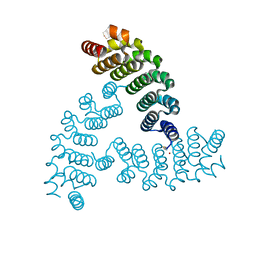 | |
6WL6
 
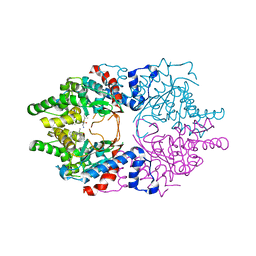 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(2R)-1-AMINO-4-METHYLPENTAN-2-YL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(2R)-1-amino-4-methylpentan-2-yl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
6VI0
 
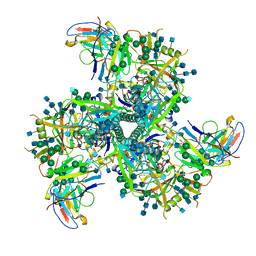 | | Cryo-EM structure of VRC01.23 in complex with HIV-1 Env BG505 DS.SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-13 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | A matrix of structure-based designs yields improved VRC01-class antibodies for HIV-1 therapy and prevention.
Mabs, 13
|
|
5Y2X
 
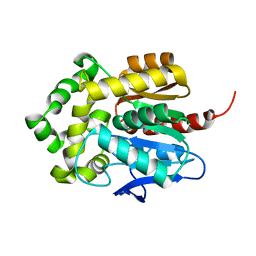 | | Crystal structure of apo-HaloTag (M175C) | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
2Z9B
 
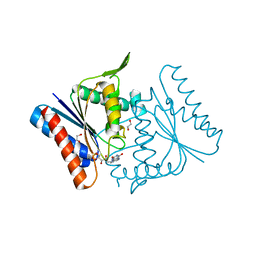 | |
7CO3
 
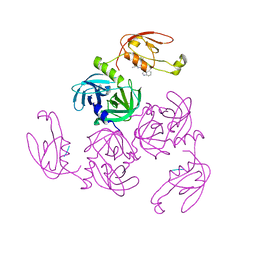 | | HtrA-type protease AlgWS227A with tripeptide | | Descriptor: | AlgW protein, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO5
 
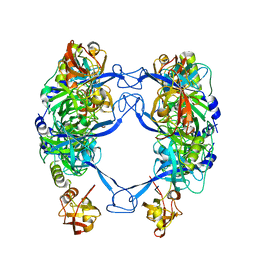 | | HtrA-type protease AlgW with decapeptide | | Descriptor: | AlgW protein, IMIDAZOLE, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO2
 
 | | HtrA-type protease AlgW with tripeptide | | Descriptor: | AlgW protein, IMIDAZOLE, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO7
 
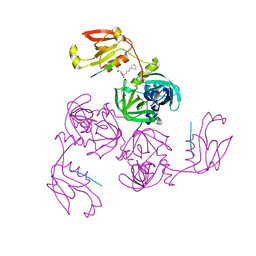 | | HtrA-type protease AlgWS227A with decapeptide | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, AlgW protein, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
8D21
 
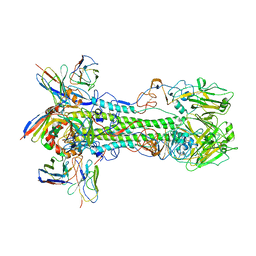 | | Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer | | Descriptor: | 1B06 Heavy Chain, 1B06 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans.
Sci Transl Med, 15, 2023
|
|
6VFI
 
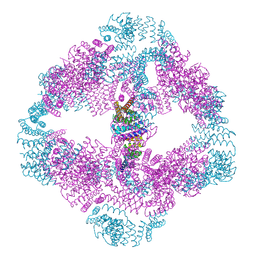 | |
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
2Z98
 
 | |
2Z9C
 
 | |
2Z9D
 
 | |
2D5I
 
 | |
5WCA
 
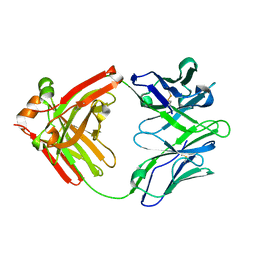 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 27-1C08 Fab. | | Descriptor: | VRC315 27-1C08 Fab Heavy chain, VRC315 27-1C08 Fab Light chain | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
5YLO
 
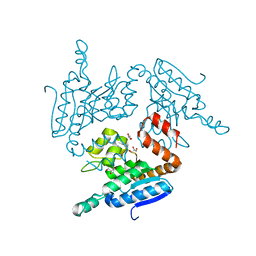 | | Structural of Pseudomonas aeruginosa PA4980 | | Descriptor: | GLYCEROL, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Li, T, Peng, C.T, Li, C.C, Xiao, Q.J, He, L.H, Wang, N.Y, Bao, R. | | Deposit date: | 2017-10-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural characterization of a Delta3, Delta2-enoyl-CoA isomerase from Pseudomonas aeruginosa: implications for its involvement in unsaturated fatty acid metabolism.
J.Biomol.Struct.Dyn., 37, 2019
|
|
1EHL
 
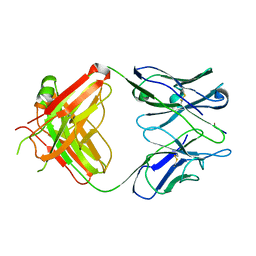 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | Descriptor: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
6JAU
 
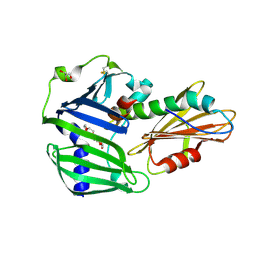 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
5WCD
 
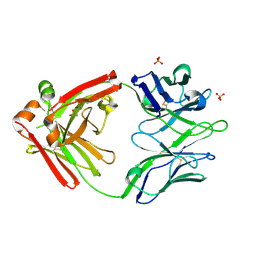 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 04-1D02 Fab. | | Descriptor: | PHOSPHATE ION, SULFATE ION, VRC315 04-1D02 Fab Heavy chain, ... | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-23 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
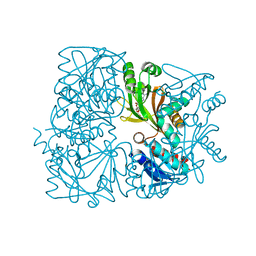
5ZK6
 
 | |
4GPL
 
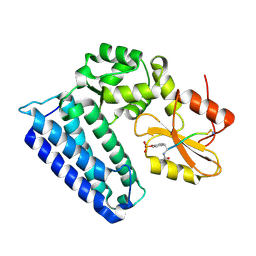 | |
