4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
6L56
 
 | | Fe(II) loaded Tegillarca granosa ferritin | | Descriptor: | FE (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85300577 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
6L55
 
 | | Recombinant Tegillarca granosa ferritin | | Descriptor: | FE (III) ION, FORMIC ACID, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78304863 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
6KZY
 
 | | Cu(II) loaded Tegillarca granosa ferritin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-09-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.30057073 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
5FVF
 
 | | Room temperature structure of IrisFP determined by serial femtosecond crystallography. | | Descriptor: | AMMONIUM ION, Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
6L58
 
 | | Cu(II) loaded Tegillarca granosa M-ferritin soaked with Fe(II) | | Descriptor: | COPPER (II) ION, Ferritin | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.90270972 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
5FVG
 
 | | Structure of IrisFP at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett, 7, 2016
|
|
4HBM
 
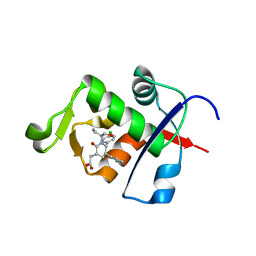 | | Ordering of the N Terminus of Human MDM2 by Small Molecule Inhibitors | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S)-1-hydroxybutan-2-yl]-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ordering of the N-terminus of human MDM2 by small molecule inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
5VR4
 
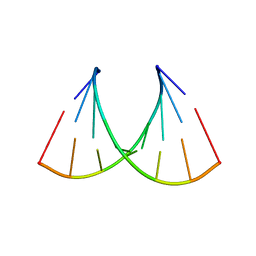 | | RNA octamer containing 2'-F-4'-OMe U. | | Descriptor: | COBALT TETRAAMMINE ION, RNA (5'-R(*CP*GP*AP*AP*(UMO)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing.
J. Am. Chem. Soc., 139, 2017
|
|
5FVI
 
 | | Structure of IrisFP in mineral grease at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
4JWR
 
 | |
7E6G
 
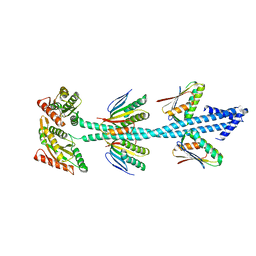 | | Crystal structure of diguanylate cyclase SiaD in complex with its activator SiaC from Pseudomonas aeruginosa | | Descriptor: | DUF1987 domain-containing protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for diguanylate cyclase activation by its binding partner in Pseudomonas aeruginosa .
Elife, 10, 2021
|
|
7E9Q
 
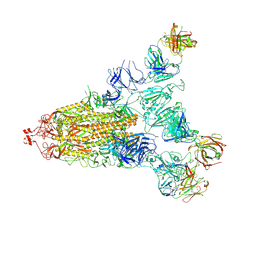 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9O
 
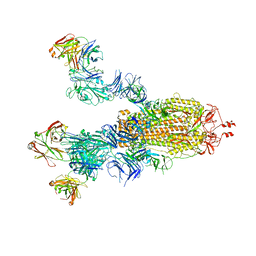 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9P
 
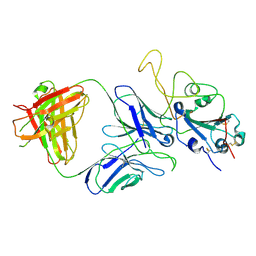 | |
7E9N
 
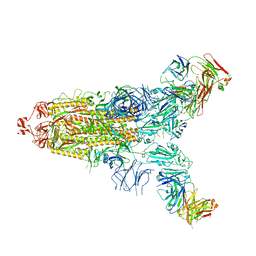 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENG
 
 | |
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
2VU4
 
 | | Structure of PsbP protein from Spinacia oleracea at 1.98 A resolution | | Descriptor: | OXYGEN-EVOLVING ENHANCER PROTEIN 2, ZINC ION | | Authors: | Lapkouski, M, Ristvejova, R, Arellano, J.B, Revuelta, J.L, Kuta Smatanova, I, Ettrich, R. | | Deposit date: | 2008-05-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Raman Spectroscopy Adds Complementary Detail to the High-Resolution X-Ray Crystal Structure of Photosynthetic Psbp from Spinacia Oleracea.
Plos One, 7, 2012
|
|
7DUU
 
 | | Crystal structure of HLA molecule with an KIR receptor | | Descriptor: | Beta-2-microglobulin, Killer cell immunoglobulin-like receptor 2DS2, LEU-ASN-PRO-SER-VAL-ALA-ALA-THR-LEU, ... | | Authors: | Yang, Y, Yin, L. | | Deposit date: | 2021-01-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Activating receptor KIR2DS2 bound to HLA-C1 reveals the novel recognition features of activating receptor.
Immunology, 165, 2022
|
|
