1XPN
 
 | | NMR structure of P. aeruginosa protein PA1324: Northeast Structural Genomics Consortium target PaP1 | | Descriptor: | hypothetical protein PA1324 | | Authors: | Cort, J.R, Ni, S, Lockert, E.E, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170).
Protein Sci., 18, 2009
|
|
1NLS
 
 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | Deposit date: | 1997-01-28 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1XSV
 
 | | X-ray crystal structure of conserved hypothetical UPF0122 protein SAV1236 from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | Hypothetical UPF0122 protein SAV1236 | | Authors: | Walker, J.R, Xu, X, Virag, C, McDonald, M.-L, Houston, S, Buzadzija, K, Vedadi, M, Dharamsi, A, Fiebig, K.M, Savchenko, A. | | Deposit date: | 2004-10-20 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
To be Published
|
|
1XJS
 
 | | Solution structure of Iron-Sulfur cluster assembly protein IscU from Bacillus subtilis, with Zinc bound at the active site. Northeast Structural Genomics Consortium Target SR17 | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Iron-Sulfur cluster assembly protein IscU
from Bacillus subtilis, with Zinc bound at the active site.
Northeast Structural Genomics Consortium Target SR17.
To be Published
|
|
1XHY
 
 | | X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor, SULFATE ION | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1XQH
 
 | | Crystal structure of a ternary complex of the methyltransferase SET9 (also known as SET7/9) with a P53 peptide and SAH | | Descriptor: | 9-mer peptide from tumor protein p53, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Chuikov, S, Kurash, J.K, Wilson, J.R, Xiao, B, Justin, N, Ivanov, G.S, McKinney, K, Tempst, P, Prives, C, Gamblin, S.J, Barlev, N.A, Reinberg, D. | | Deposit date: | 2004-10-12 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Regulation of p53 activity through lysine methylation
Nature, 432, 2004
|
|
1Y6L
 
 | | Human ubiquitin conjugating enzyme E2E2 | | Descriptor: | Ubiquitin-conjugating enzyme E2E2 | | Authors: | Walker, J.R, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1Y28
 
 | | Crystal structure of the R220A metBJFIXL HEME domain | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sensor protein fixL | | Authors: | Dunham, C.M, Dioum, E.M, Tuckerman, J.R, Gonzalez, G, Scott, W.G, Gilles-Gonzalez, M.A. | | Deposit date: | 2004-11-21 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A distal arginine in the oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure
Biochemistry, 42, 2003
|
|
1U11
 
 | | PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) from the acidophile Acetobacter aceti | | Descriptor: | CITRIC ACID, PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) | | Authors: | Settembre, E.C, Chittuluru, J.R, Mill, C.P, Kappock, T.J, Ealick, S.E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acidophilic adaptations in the structure of Acetobacter aceti N5-carboxyaminoimidazole ribonucleotide mutase (PurE).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2Z
 
 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
1TSH
 
 | |
1Y7Q
 
 | | Mammalian SCAN domain dimer is a domain-swapped homologue of the HIV capsid C-terminal domain | | Descriptor: | Zinc finger protein 174 | | Authors: | Ivanov, D, Stone, J.R, Maki, J.L, Collins, T, Wagner, G. | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mammalian SCAN Domain Dimer Is a Domain-Swapped Homolog of the HIV Capsid C-Terminal Domain
Mol.Cell, 17, 2005
|
|
1YD9
 
 | | 1.6A Crystal Structure of the Non-Histone Domain of the Histone Variant MacroH2A1.1. | | Descriptor: | Core histone macro-H2A.1, GOLD ION | | Authors: | Chakravarthy, S, Swamy, G.Y.S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the histone variant macroH2A
Mol.Cell.Biol., 25, 2005
|
|
3N3K
 
 | | The catalytic domain of USP8 in complex with a USP8 specific inhibitor | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 8, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Allali-Hassani, A, Lam, R, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
2HZ8
 
 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
2NQ3
 
 | | Crystal structure of the C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase | | Descriptor: | CHLORIDE ION, Itchy homolog E3 ubiquitin protein ligase | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase
To be Published
|
|
2NSQ
 
 | | Crystal structure of the C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase NEDD4-like protein, GLYCEROL | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein
To be Published
|
|
2AOT
 
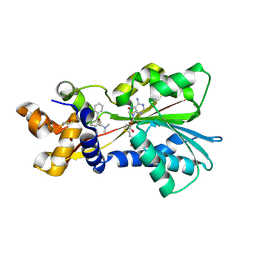 | | Histamine Methyltransferase Complexed with the Antihistamine Drug Diphenhydramine | | Descriptor: | Histamine N-methyltransferase, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
1YPN
 
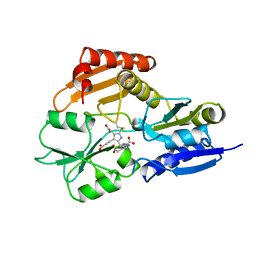 | | REDUCED FORM HYDROXYMETHYLBILANE SYNTHASE (K59Q MUTANT) CRYSTAL STRUCTURE AFTER 2 HOURS IN A FLOW CELL DETERMINED BY TIME-RESOLVED LAUE DIFFRACTION | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 1998-06-26 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-Resolved Structures of Hydroxymethylbilane Synthase (Lys59Gln Mutant) as It Isloaded with Substrate in the Crystal Determined by Laue Diffraction
J.Chem.Soc.,Faraday Trans., 94, 1998
|
|
1YFL
 
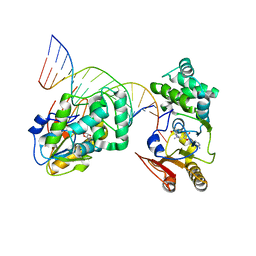 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | Descriptor: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
2BT4
 
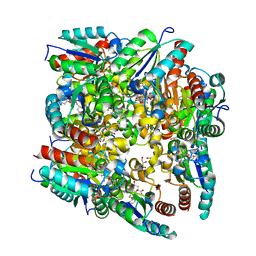 | | Type II Dehydroquinase inhibitor complex | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Toscano, M.D, Stewart, K.A, Coggins, J.R, Lapthorn, A.J, Abell, C. | | Deposit date: | 2005-05-26 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design of New Bifunctional Inhibitors of Type II Dehydroquinase.
Org.Biomol.Chem., 3, 2005
|
|
2AOW
 
 | | Histamine Methyltransferase (Natural Variant I105) Complexed with the Acetylcholinesterase Inhibitor and Altzheimer's Disease Drug Tacrine | | Descriptor: | Histamine N-methyltransferase, TACRINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
2AOX
 
 | | Histamine Methyltransferase (Primary Variant T105) Complexed with the Acetylcholinesterase Inhibitor and Altzheimer's Disease Drug Tacrine | | Descriptor: | Histamine N-methyltransferase, TACRINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
2AOV
 
 | | Histamine Methyltransferase Complexed with the Antifolate Drug Metoprine | | Descriptor: | 4-(DIMETHYLAMINO)BUTYL IMIDOTHIOCARBAMATE, 5-(3,4-DICHLOROPHENYL)-6-METHYLPYRIMIDINE-2,4-DIAMINE, Histamine N-methyltransferase | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
2CIJ
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) with bound methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Barinka, C, Plechanovova, A, Rulisek, L, Mlcochova, P, Majer, P, Slusher, B.S, Hilgenfeld, R, Mesters, J.R, Konvalinka, J. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutamate Carboxypeptidase II
Handbook of Metalloproteins, 4, 2011
|
|
