6KUY
 
 | | Crystal structure of the alpha2A adrenergic receptor in complex with a partial agonist | | Descriptor: | (2~{S})-4-fluoranyl-2-(1~{H}-imidazol-5-yl)-1-propan-2-yl-2,3-dihydroindole, Alpha2A adrenergic receptor, DI(HYDROXYETHYL)ETHER | | Authors: | Qu, L, Zhou, Q.T, Wu, D, Zhao, S.W. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the alpha2A adrenergic receptor in complex with a partial agonist
To Be Published
|
|
6X6F
 
 | | The structure of Pf6r from the filamentous phage Pf6 of Pseudomonas aeruginosa PA01 | | Descriptor: | NITRATE ION, Pf6r | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
6U7K
 
 | | Prefusion structure of PEDV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epidemic Diarrhea Virus Spike Protein in the Prefusion Conformation.
J.Virol., 93, 2019
|
|
2RKN
 
 | | X-ray structure of the self-defense and signaling protein DIR1 from Arabidopsis taliana | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, DIR1 protein, ZINC ION | | Authors: | Lascombe, M.B, Prange, T, Buhot, N, Marion, D, Bakan, B, Lamb, C. | | Deposit date: | 2007-10-17 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of "defective in induced resistance" protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein.
Protein Sci., 17, 2008
|
|
5AZ6
 
 | |
6KYB
 
 | | Crystal structure of Atg18 from Saccharomyces cerevisiae | | Descriptor: | Autophagy-related protein 18 | | Authors: | Tang, D, Lei, Y, Liao, G, Chen, Q, Xu, L, Lu, K, Qi, S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of Atg18 reveals a new binding site for Atg2 in Saccharomyces cerevisiae.
Cell.Mol.Life Sci., 78, 2021
|
|
5B2D
 
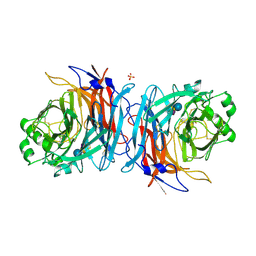 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to 3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6U5F
 
 | | CryoEM Structure of Pyocin R2 - precontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622, Tube PA0623 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6KXF
 
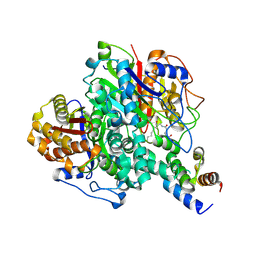 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
1O2A
 
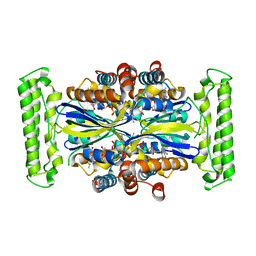 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD at 1.8 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
6L1R
 
 | |
6XKD
 
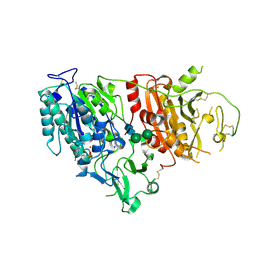 | | Structure of ligand-bound mouse cGAMP hydrolase ENPP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Li, L. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Aided Development of Small-Molecule Inhibitors of ENPP1, the Extracellular Phosphodiesterase of the Immunotransmitter cGAMP.
Cell Chem Biol, 27, 2020
|
|
2SBA
 
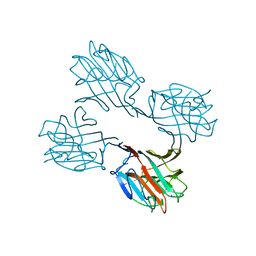 | | SOYBEAN AGGLUTININ COMPLEXED WITH 2,6-PENTASACCHARIDE | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Dessen, A, Gupta, D, Sabesan, S, Brewer, C.F, Sacchettini, J.C. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of the soybean agglutinin cross-linked with a biantennary analog of the blood group I carbohydrate antigen.
Biochemistry, 34, 1995
|
|
6U60
 
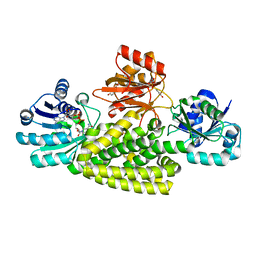 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | Authors: | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
5BSK
 
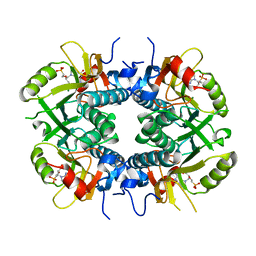 | | Human HGPRT in complex with (S)-HPEPG, an acyclic nucleoside phosphonate | | Descriptor: | (2-{[(2S)-1-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3-hydroxypropan-2-yl]oxy}ethyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Keough, D.T, Guddat, L.W, Kaiser, M.M, Hockova, D, Wang, T.-H, Janeba, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synthesis and Evaluation of Novel Acyclic Nucleoside Phosphonates as Inhibitors of Plasmodium falciparum and Human 6-Oxopurine Phosphoribosyltransferases.
Chemmedchem, 10, 2015
|
|
5BT5
 
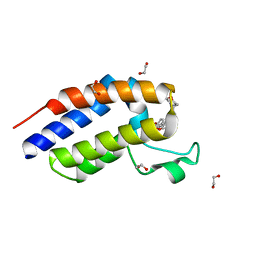 | | Crystal structure of BRD2 second bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 2 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BRD2 second bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5BMX
 
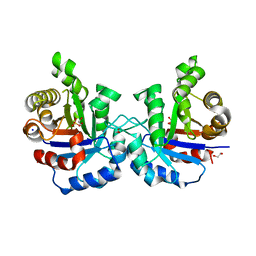 | | Crystal structure of T75N mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
6LB5
 
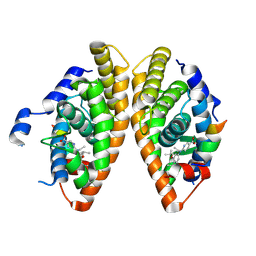 | | Crystal structure of dimeric RXR-LBD complexed with full agonist NEt-3IB and TIF2 co-activator | | Descriptor: | 6-[ethyl-[3-(2-methylpropoxy)-4-propan-2-yl-phenyl]amino]pyridine-3-carboxylic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Imai, D, Numoto, N, Nakano, S, Kakuta, H, Ito, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric RXR-LBD complexed with full agonist NEt-3IB and TIF2 co-activator
To Be Published
|
|
5C7C
 
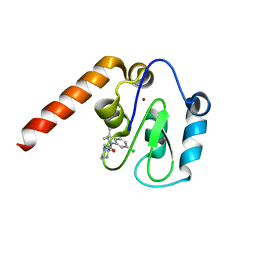 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 18 | | Descriptor: | (2R)-4-[2-(6-chloro-3,3-dimethyl-2,3-dihydro-1H-indol-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6LHY
 
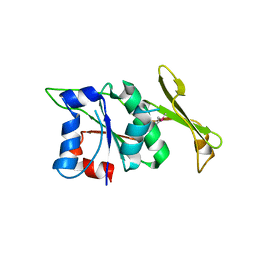 | | Crystal structure of ThsB | | Descriptor: | DUF1863 domain-containing protein | | Authors: | Bae, E, Ka, D, Oh, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural and functional evidence of bacterial antiphage protection by Thoeris defense system via NAD+degradation.
Nat Commun, 11, 2020
|
|
5BZX
 
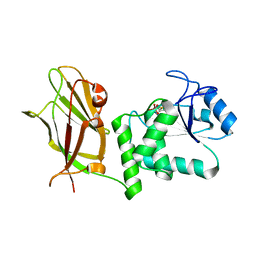 | | Crystal structure of human phosphatase PTEN treated with a bisperoxovanadium complex | | Descriptor: | L(+)-TARTARIC ACID, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, VANADATE ION | | Authors: | Lee, C.-U, Bier, D, Hennig, S, Grossmann, T.N. | | Deposit date: | 2015-06-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redox Modulation of PTEN Phosphatase Activity by Hydrogen Peroxide and Bisperoxidovanadium Complexes.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5CBU
 
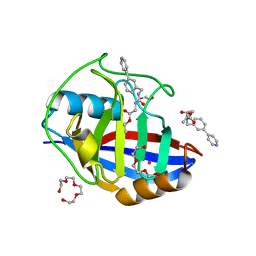 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
5CC9
 
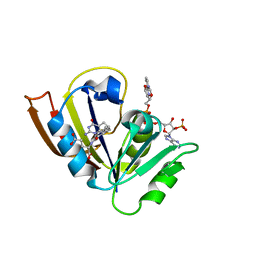 | |
6XLC
 
 | | Full-length Hsc82 bound to AMPPNP | | Descriptor: | ATP-dependent molecular chaperone HSC82, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Full-length Hsc82 bound to AMPPNP
To Be Published
|
|
6KXD
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
