7WWR
 
 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
7XAN
 
 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-03-18 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
7WWS
 
 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-05-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7X2E
 
 | |
4HU8
 
 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
4J0U
 
 | | Crystal structure of IFIT5/ISG58 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5 | | Authors: | Liu, Y, Liang, H, Feng, F, Yuan, L, Wang, Y.E, Crowley, C, Lv, Z, Li, J, Zeng, S, Cheng, G. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Crystal Structure of IFIT5
To be Published
|
|
4HY6
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1 | | Descriptor: | 6,6-dimethyl-3-(trifluoromethyl)-1,5,6,7-tetrahydro-4H-indazol-4-one, Heat shock protein HSP 90-alpha | | Authors: | Yang, M, Li, J, He, J.H. | | Deposit date: | 2012-11-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1
to be published
|
|
8XOY
 
 | | The Crystal Structure of PTP1B from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Li, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of PTP1B from Biortus.
To Be Published
|
|
8WWS
 
 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
8XI3
 
 | | Structure of mouse SCMC-14-3-3gama complex | | Descriptor: | 14-3-3 protein gamma, NACHT, LRR and PYD domains-containing protein 5, ... | | Authors: | Chi, P, Han, Z, Jiao, H, Li, J, Wang, X, Hu, H, Deng, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of mouse SCMC-14-3-3gama complex
To Be Published
|
|
6LAR
 
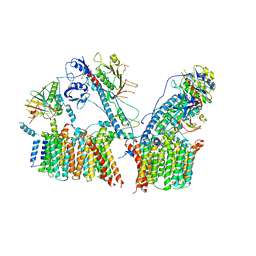 | | Structure of ESX-3 complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Wang, S.H, Zhou, K.X, Li, J, Rao, Z.H. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | cryo-em structure of esx-3
To Be Published
|
|
7VBD
 
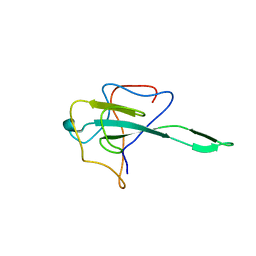 | | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0 | | Descriptor: | Nucleoprotein | | Authors: | Zeng, P, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-08-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0
To Be Published
|
|
7VBF
 
 | |
7VBE
 
 | |
7XB3
 
 | |
8K4Y
 
 | | Structure of a triple-helix region of human ReCol 3 from Trautec | | Descriptor: | SULFATE ION, triple-helix region of human ReCol 3 | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human ReCol 3 from Trautec.
To Be Published
|
|
8K4X
 
 | | Structure of a triple-helix region of human Collagen type XVII from Trautec | | Descriptor: | triple-helix region of human Collagen type XVII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type XVII from Trautec.
To Be Published
|
|
8K4W
 
 | | Structure of a triple-helix region of human collagen type I from Trautec | | Descriptor: | CHLORIDE ION, triple-helix region of human collagen type I | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human collagen type I from Trautec.
To Be Published
|
|
1B34
 
 | | CRYSTAL STRUCTURE OF THE D1D2 SUB-COMPLEX FROM THE HUMAN SNRNP CORE DOMAIN | | Descriptor: | PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D1), PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D2) | | Authors: | Walke, S, Young, R.J, Kambach, C, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-17 | | Release date: | 2000-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
5X61
 
 | | Crystal structure of Acetylcholinesterase Catalytic Subunit of the Malaria Vector Anopheles Gambiae, 3.4 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Han, Q, Robinson, H, Ding, H, Wong, D.M, Lam, P.C.H, Totrov, M.M, Carlier, P.R, Li, J. | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae
Insect Sci., 2017
|
|
5YDJ
 
 | | Crystal structure of anopheles gambiae acetylcholinesterase in complex with PMSF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Han, Q, Guan, H, Ding, H, Liao, C, Robinson, H, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5Y4F
 
 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | Descriptor: | ACETATE ION, Ankyrin-2, CALCIUM ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
