4OEW
 
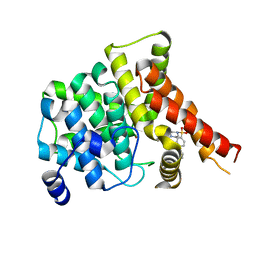 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-iodo-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
4OEX
 
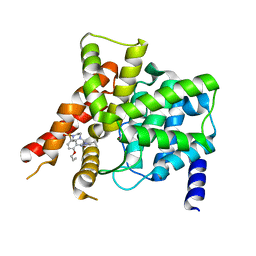 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
9IK2
 
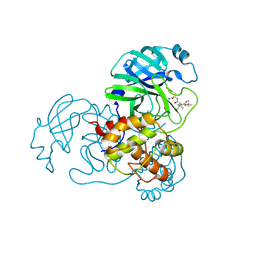 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
7X31
 
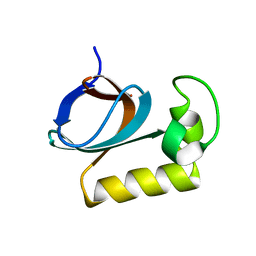 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
6LRM
 
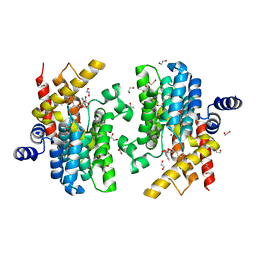 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6LRD
 
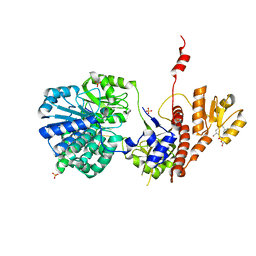 | | Structure of RecJ complexed with a 5'-P-dSpacer-modified ssDNA | | Descriptor: | ASP-LEU-PRO-PHE, DNA (5'-D(P*(3DR)P*TP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Cheng, K, Hua, Y. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901335 Å) | | Cite: | Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans.
Nucleic Acids Res., 48, 2020
|
|
4I9U
 
 | | Crystal structure of rabbit LDHA in complex with a fragment inhibitor AP26256 | | Descriptor: | 6-({2-[(5-chloro-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
6KZD
 
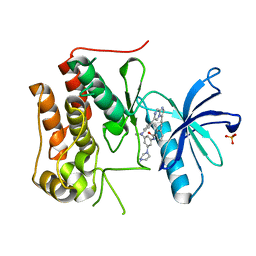 | | Crystal structure of TRKc in complex with 3-((6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl)ethynyl)- N-(3-isopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-2- methylbenzamide | | Descriptor: | 3-[2-[6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl]ethynyl]-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-propan-2-yl-phenyl]benzamide, NT-3 growth factor receptor, PHOSPHATE ION | | Authors: | Wang, Y, Zhang, Z.M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
8DHL
 
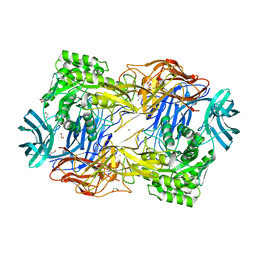 | | Tannerella forsythia beta-glucuronidase (L2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHW
 
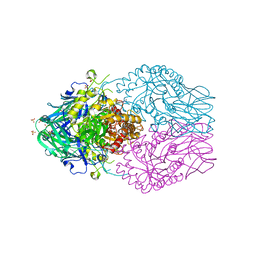 | | Treponema lecithinolyticum beta-glucuronidase in complex with a UNC4917-glucuronide conjugate | | Descriptor: | 4-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-2,7-bis(methylamino)pyrido[3',2':4,5]thieno[3,2-d]pyrimidine, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHV
 
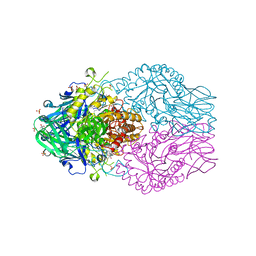 | | Treponema lecithinolyticum beta-glucuronidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHE
 
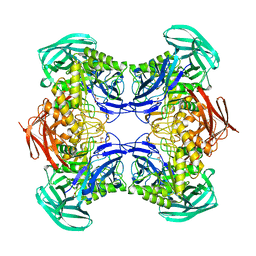 | |
7X4B
 
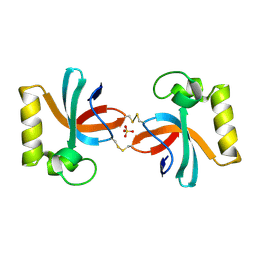 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
8CQ2
 
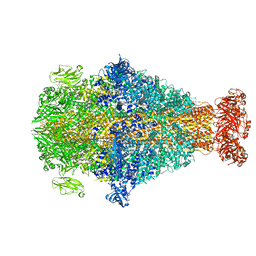 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
8CPZ
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K1179W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
8CQ0
 
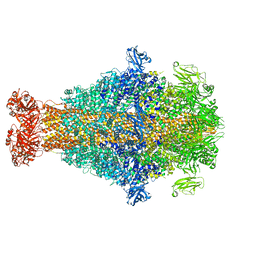 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
8DKE
 
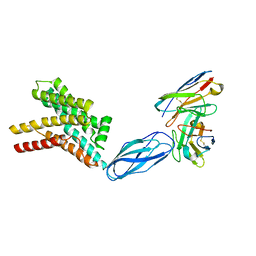 | |
8DKW
 
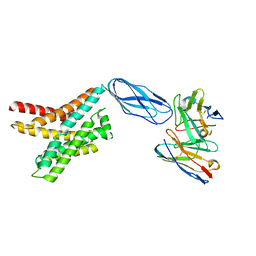 | |
8DKI
 
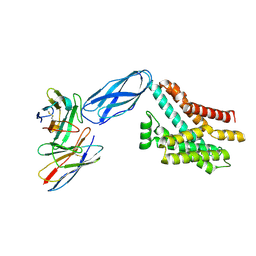 | |
8DKX
 
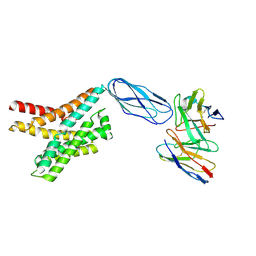 | |
8DKM
 
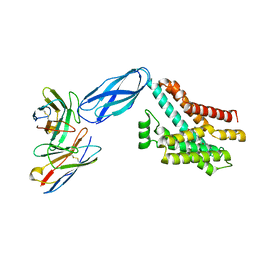 | |
4RLW
 
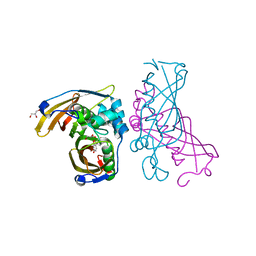 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Butein | | Descriptor: | (2E)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)prop-2-en-1-one, (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
8DYP
 
 | |
4RLU
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with 2',4,4'-trihydroxychalcone | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 2',4,4'-TRIHYDROXYCHALCONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
