6YWK
 
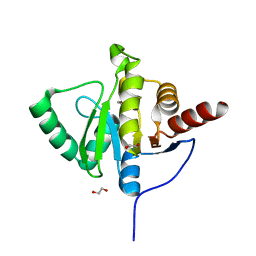 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6GL9
 
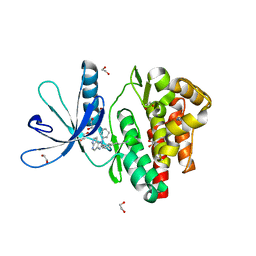 | | Crystal structure of JAK3 in complex with Compound 10 (FM475) | | Descriptor: | (~{E})-3-[3-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)phenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
3V2O
 
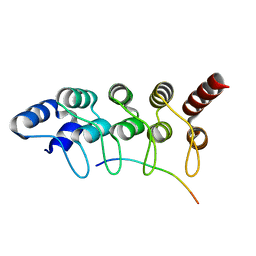 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human ANKRA2 | | Descriptor: | Ankyrin repeat family A protein 2, Low-density lipoprotein receptor-related protein 2 | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
3V31
 
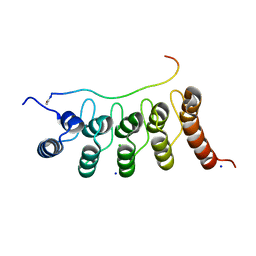 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human ANKRA2 | | Descriptor: | Ankyrin repeat family A protein 2, CHLORIDE ION, Histone deacetylase 4, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
3V2X
 
 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human ANKRA2 | | Descriptor: | Ankyrin repeat family A protein 2, Low-density lipoprotein receptor-related protein 2 | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
3V30
 
 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human RFXANK | | Descriptor: | DNA-binding protein RFX5, DNA-binding protein RFXANK | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
6GLA
 
 | | Crystal structure of JAK3 in complex with Compound 11 (FM481) | | Descriptor: | (~{E})-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)furan-2-yl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
7ZHQ
 
 | | Crystal structure of TTBK1 in complex with compound 10 (7-009) | | Descriptor: | (3~{S})-1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-methyl-pent-1-yn-3-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHO
 
 | | Crystal structure of TTBK1 in complex with compound 3 (7-001) | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(2-azanylpyrimidin-4-yl)-1~{H}-indol-5-yl]-2-methyl-but-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHP
 
 | | Crystal structure of TTBK1 in complex with compound 9 (7-005) | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-ethyl-pent-1-yn-3-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
6MLC
 
 | | PHD6 domain of MLL3 in complex with histone H4 | | Descriptor: | GLYCEROL, Histone H4, Histone-lysine N-methyltransferase 2C, ... | | Authors: | Dong, A, Liu, Y, Qin, S, Lei, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4.
Nat Commun, 10, 2019
|
|
4Y03
 
 | | Crystal Structure of the fifth bromodomain of human PB1 in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, CITRIC ACID, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of the fifth bromodomain of human PB1 in complex with salicylic acid
To Be Published
|
|
4XY9
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
4XYA
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-1-benzofuran-7-yl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
4XUB
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with BAZ2-ICR chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-(1-methyl-1H-pyrazol-4-yl)-1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
4XY8
 
 | | Crystal Structure of the bromodomain of BRD9 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, BROMIDE ION, Bromodomain-containing protein 9 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
6X9O
 
 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
8BI8
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase | | Descriptor: | Nitric oxide synthase, brain | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
8BI9
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase (T112W/T116E variant) | | Descriptor: | Nitric oxide synthase, brain, TERTIARY-BUTYL ALCOHOL | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
8A31
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC694 | | Descriptor: | 4-(3-fluoranylpyrrol-1-yl)-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8A32
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC769 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3,4-bis(fluoranyl)pyrrol-1-yl]-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
3ZZW
 
 | | Crystal structure of the kinase domain of ROR2 | | Descriptor: | CHLORIDE ION, SULFATE ION, TYROSINE-PROTEIN KINASE TRANSMEMBRANE RECEPTOR ROR2 | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Karlberg, T, Nyman, T, Schuler, H, Thorsell, A.G, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-05 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Kinase Domain of Ror2
To be Published
|
|
3UOW
 
 | | Crystal Structure of PF10_0123, a GMP Synthetase from Plasmodium Falciparum | | Descriptor: | CALCIUM ION, GMP synthetase, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Wernimont, A.K, Dong, A, Hills, T, Amani, M, Perieteanu, A, Lin, Y.H, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structure of PF10_0123, a GMP Synthetase from Plasmodium Falciparum
TO BE PUBLISHED
|
|
5KH9
 
 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | Authors: | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
