7D39
 
 | | FLR-apo | | Descriptor: | Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D3B
 
 | | flavone reductase | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D3A
 
 | | flavone reductase | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D38
 
 | | flavone reductase | | Descriptor: | Cd1, FLAVIN MONONUCLEOTIDE, chrysin | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
4N0S
 
 | | Complex of ERK2 with caffeic acid | | Descriptor: | CAFFEIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2013-10-02 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7992 Å) | | Cite: | Caffeic Acid Directly Targets ERK1/2 to Attenuate Solar UV-Induced Skin Carcinogenesis.
Cancer Prev Res (Phila), 7, 2014
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
8IOY
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | Descriptor: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yang, G, Xu, L, Guo, J, Wu, Z. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7DRI
 
 | | Structure of SspE_CTD_41658 | | Descriptor: | DUF1524 domain | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRS
 
 | | Structure of SspE_40224 | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
 | | Structure of SspE-R100A protein | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7XUK
 
 | |
7XUM
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with Cu+ in presence of ATOX1 | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 2 | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7XUN
 
 | |
7XUO
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with cisplatin in presence of ATOX1 | | Descriptor: | Copper-transporting ATPase 2, PLATINUM (II) ION | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
5XLS
 
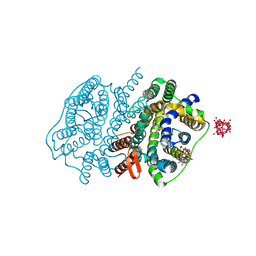 | | Crystal structure of UraA in occluded conformation | | Descriptor: | 12-TUNGSTOPHOSPHATE, URACIL, Uracil permease | | Authors: | Yu, X.Z, Yang, G.H, Yan, C.Y, Yan, N. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters
Cell Res., 27, 2017
|
|
6KVA
 
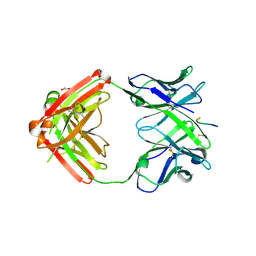 | | Structure of anti-hCXCR2 abN48-2 in complex with its CXCR2 epitope | | Descriptor: | 1,2-ETHANEDIOL, Peptide from C-X-C chemokine receptor type 2, heavy chain, ... | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
6KVF
 
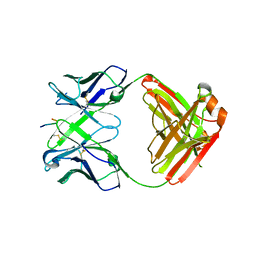 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
5FA5
 
 | |
7CWO
 
 | | SARS-CoV-2 spike protein RBD and P17 fab complex | | Descriptor: | Spike glycoprotein, heavy chain of P17 Fab, light chain of P17 Fab | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
7CWL
 
 | | SARS-CoV-2 spike protein and P17 fab complex with one RBD in close state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab P17 heavy chain, Fab P17 light chain, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
7CWM
 
 | |
7CWN
 
 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
6JYZ
 
 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
8X7X
 
 | | Crystal structure of SADS-CoV fusion core | | Descriptor: | CHLORIDE ION, HR1, HR2 | | Authors: | Yan, L. | | Deposit date: | 2023-11-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Fusion Cores from CCoV-HuPn-2018 and SADS-CoV.
Viruses, 16, 2024
|
|
5ZT3
 
 | | Crystal structure of WA352 from Oryza sativa | | Descriptor: | WA352 | | Authors: | Wang, X, Guan, Z, Yin, P. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Crystal structure of WA352 provides insight into cytoplasmic male sterility in rice
Biochem. Biophys. Res. Commun., 501, 2018
|
|
