5XDE
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDG
 
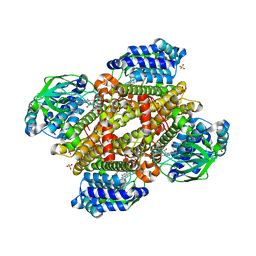 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene sulfoxide | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
1S0M
 
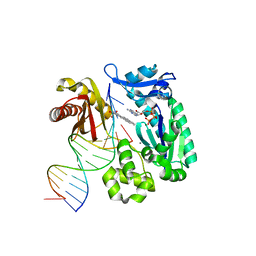 | | Crystal structure of a Benzo[a]pyrene Diol Epoxide adduct in a ternary complex with a DNA polymerase | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*T)-3', ... | | Authors: | Ling, H, Sayer, J.M, Boudsocq, F, Plosky, B.S, Woodgate, R, Yang, W. | | Deposit date: | 2003-12-31 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2NLI
 
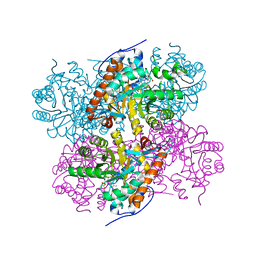 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
2ZFA
 
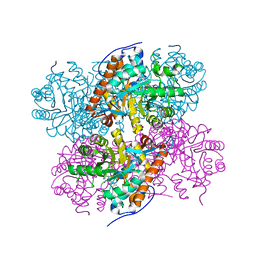 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
3APA
 
 | | Crystal structure of human pancreatic secretory protein ZG16p | | Descriptor: | CHLORIDE ION, GLYCEROL, Zymogen granule membrane protein 16 | | Authors: | Kanagawa, M, Satoh, T, Nakano, Y, Kojima-Aikawa, K, Yamaguchi, Y. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related beta-prism fold
Biochem.Biophys.Res.Commun., 2010
|
|
3AQG
 
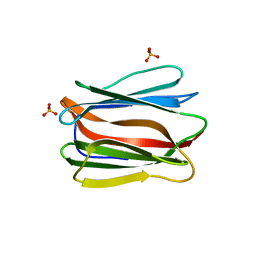 | | Crystal structure of human pancreatic secretory protein ZG16b | | Descriptor: | PHOSPHATE ION, Zymogen granule protein 16 homolog B | | Authors: | Kanagawa, M, Satoh, T, Nakano, Y, Kojima-Aikawa, K, Yamaguchi, Y. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related beta-prism fold
Biochem.Biophys.Res.Commun., 2010
|
|
5GZ8
 
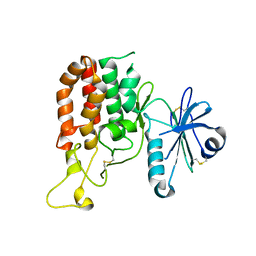 | |
5GZ9
 
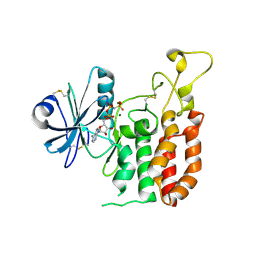 | | Crystal structure of catalytic domain of Protein O-mannosyl Kinase in complexes with AMP-PNP, Magnesium ions and glycopeptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein O-mannose kinase, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2016-09-27 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy.
Genes Cells, 22, 2017
|
|
5DSV
 
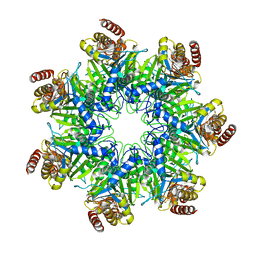 | | Crystal structure of human proteasome alpha7 tetradecamer | | Descriptor: | Proteasome subunit alpha type-3 | | Authors: | Satoh, T, Thammaporn, R, Seetaha, S, Kato, K. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Disassembly of the self-assembled, double-ring structure of proteasome alpha 7 homo-tetradecamer by alpha 6
Sci Rep, 5, 2015
|
|
2LRR
 
 | | Solution structure of the R3H domain from human Smubp-2 in complex with 2'-deoxyguanosine-5'-monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-binding protein SMUBP-2 | | Authors: | Jaudzems, K, Zhulenkovs, D, Otting, G, Liepinsh, E. | | Deposit date: | 2012-04-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for 5'-End-Specific Recognition of Single-Stranded DNA by the R3H Domain from Human Smubp-2
J.Mol.Biol., 12, 2012
|
|
4GX9
 
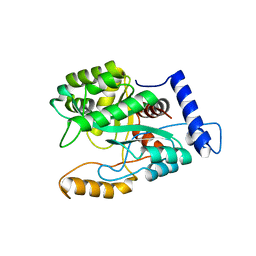 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
4GX8
 
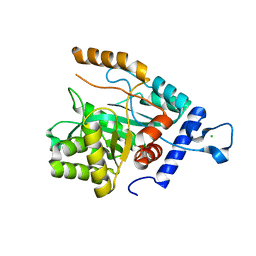 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | CHLORIDE ION, DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Robinson, A, Horan, N, Xu, Z.-Q, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
2I9G
 
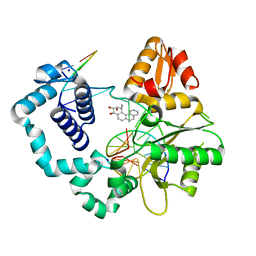 | | DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Wilson, S.H, Beard, W.A, Pedersen, L.C. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of DNA polymerase beta with a benzo[c]phenanthrene diol epoxide-adducted template exhibits mutagenic features.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5UG7
 
 | | Calcium bound Perforin C2 Domain - T431D | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
5UG6
 
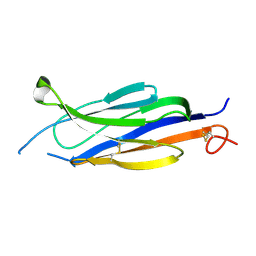 | | Perforin C2 Domain - T431D | | Descriptor: | IODIDE ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
5ETY
 
 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
7CE4
 
 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
3W56
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
3W57
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein, CALCIUM ION | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
