6HXB
 
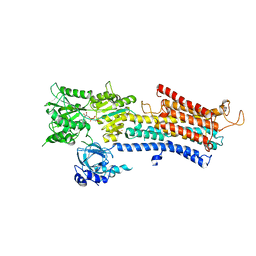 | | SERCA2a from pig heart | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Sitsel, A, Andersen, J.L, Nissen, P, Olesen, C. | | Deposit date: | 2018-10-16 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of the heart specific SERCA2a Ca 2+ -ATPase.
Embo J., 38, 2019
|
|
7BH2
 
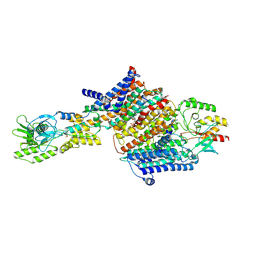 | | Cryo-EM Structure of KdpFABC in E2Pi state with BeF3 and K+ | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BH1
 
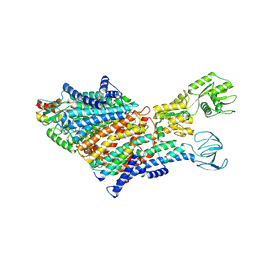 | | Cryo-EM Structure of KdpFABC in E1 state with K | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BGY
 
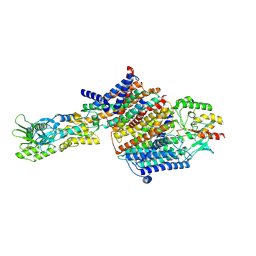 | | Cryo-EM Structure of KdpFABC in E2Pi state with MgF4 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8RM7
 
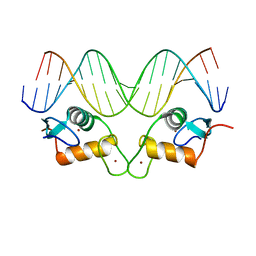 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: MMTV-177 GRE/ARE | | Descriptor: | Isoform 2 of Androgen receptor, MMTV-177 GRE/ARE Chain C, MMTV-177 GRE/ARE, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
8RM6
 
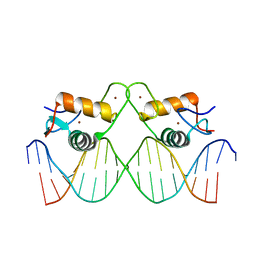 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: C3(1)ARE | | Descriptor: | C3(1)ARE_Chain C, C3(1)ARE_Chain D, Isoform 2 of Androgen receptor, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
6SAO
 
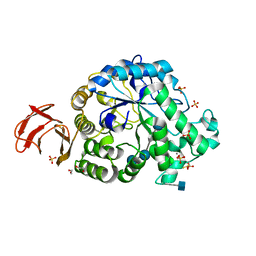 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAV
 
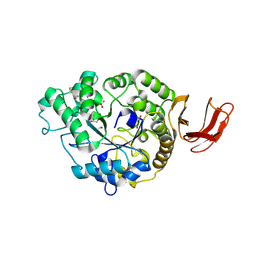 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAU
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6WJG
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
6WJF
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
4QLT
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 2 (PR924) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4QLV
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 17 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4QLS
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 11 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-D-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | De Bruin, G, Huber, E, Xin, B, Van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, Van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4U8H
 
 | | Crystal Structure of Mammalian Period-Cryptochrome Complex | | Descriptor: | Cryptochrome-2, Period circadian protein homolog 2, ZINC ION | | Authors: | Nangle, S.N, Rosensweig, C, Koike, N, Tei, H, Takahashi, J.S, Green, C.B, Zheng, N. | | Deposit date: | 2014-08-03 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex.
Elife, 3, 2014
|
|
4FGD
 
 | | Structure of the effector protein Tse1 from Pseudomonas aeruginosa, selenomethionine variant | | Descriptor: | CHLORIDE ION, THIOCYANATE ION, Tse1 | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
8Q3H
 
 | | Capra hircus reactive intermediate deaminase A mutant - A108D | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, GLYCEROL | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capra hircus reactive intermediate deaminase A mutant - A108D
To Be Published
|
|
4FGE
 
 | | Structure of the effector protein Tse1 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, THIOCYANATE ION, ... | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
4FGI
 
 | | Structure of the effector - immunity system Tse1 / Tsi1 from Pseudomonas aeruginosa | | Descriptor: | Tse1, Tsi1 | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
8QOV
 
 | | Capra hircus reactive intermediate deaminase A mutant - I126Y | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Capra hircus reactive intermediate deaminase A mutant - I126Y
To Be Published
|
|
8QOU
 
 | | Reactive intermediate deaminase A mutant - R107K | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reactive intermediate deaminase A mutant - R107K
To Be Published
|
|
8QOK
 
 | |
8QOP
 
 | |
8QOQ
 
 | |
8QOM
 
 | |
