7ZBC
 
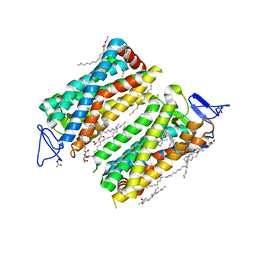 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, S, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
6QNO
 
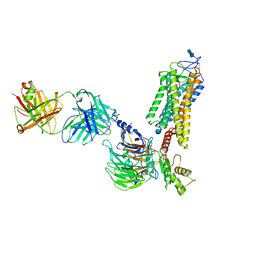 | | Rhodopsin-Gi protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab antibody fragment heavy chain, Fab antibody fragment light chain, ... | | Authors: | Tsai, C.-J, Marino, J, Adaixo, R.J, Pamula, F, Muehle, J, Maeda, S, Flock, T, Taylor, N.M.I, Mohammed, I, Matile, H, Dawson, R.J.P, Deupi, X, Stahlberg, H, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
6QNK
 
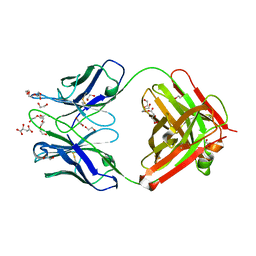 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
4BVN
 
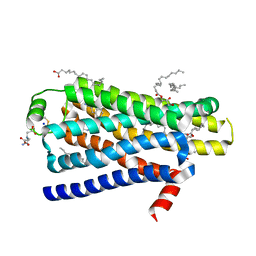 | | Ultra-thermostable beta1-adrenoceptor with cyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, ... | | Authors: | Miller, J, Nehme, R, Warne, T, Edwards, P.C, Leslie, A.G.W, Schertler, G, Tate, C.G. | | Deposit date: | 2013-06-26 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Resolution Structure of Cyanopindolol-Bound Beta1- Adrenoceptor Identifies an Intramembrane Na+ Ion that Stabilises the Ligand-Free Receptor.
Plos One, 9, 2014
|
|
8BX7
 
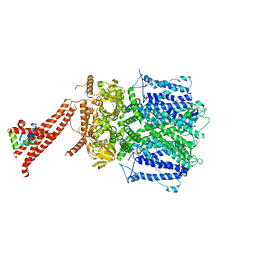 | | Structure of the rod CNG channel bound to calmodulin | | Descriptor: | Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1, calmodulin-1 | | Authors: | Marino, J. | | Deposit date: | 2022-12-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of calmodulin modulation of the rod cyclic nucleotide-gated channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4BEY
 
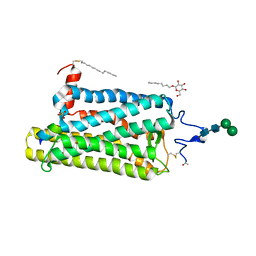 | | Night blindness causing G90D rhodopsin in complex with GaCT2 peptide | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Singhal, A, Ostermaier, M.K, Vishnivetskiy, S.A, Panneels, V, Homan, K.T, Tesmer, J.J.G, Veprintsev, D, Deupi, X, Gurevich, V.V, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin.
Embo Rep., 14, 2013
|
|
4BEZ
 
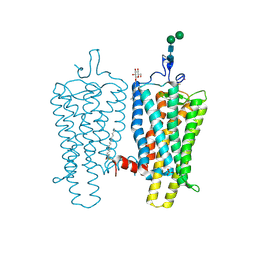 | | Night blindness causing G90D rhodopsin in the active conformation | | Descriptor: | ACETATE ION, PALMITIC ACID, RHODOPSIN, ... | | Authors: | Singhal, A, Ostermaier, M.K, Vishnivetskiy, S.A, Panneels, V, Homan, K.T, Tesmer, J.J.G, Veprintsev, D, Deupi, X, Gurevich, V.V, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin.
Embo Rep., 14, 2013
|
|
6CRK
 
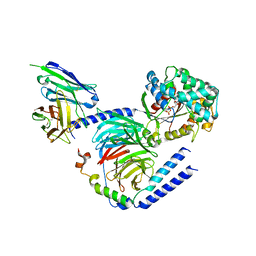 | | Heterotrimeric G-protein in complex with an antibody fragment | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Maeda, S, Dawson, R, Kobilka, B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an antibody fragment that stabilizes GPCR/G-protein complexes.
Nat Commun, 9, 2018
|
|
6DDE
 
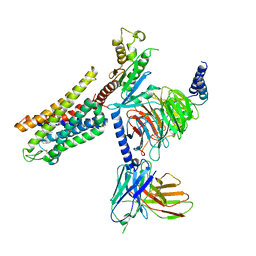 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Zhang, Y, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
6DDF
 
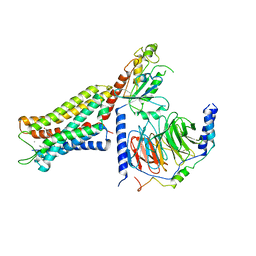 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
2YCW
 
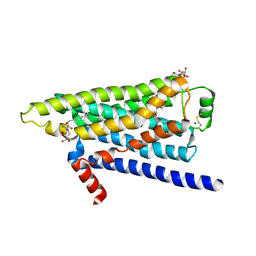 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST CARAZOLOL | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, BETA-1 ADRENERGIC RECEPTOR, HEGA-10, ... | | Authors: | Moukhametzianov, R, Warne, T, Edwards, P.C, Serrano-Vega, M.J, Leslie, A.G.W, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a Beta-1- Adrenergic Receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YCX
 
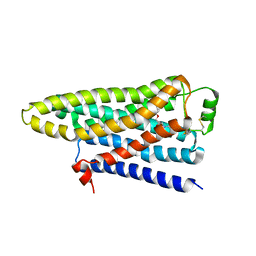 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST CYANOPINDOLOL | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, BETA-1 ADRENERGIC RECEPTOR, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Moukhametzianov, R, Warne, T, Edwards, P.C, Serrano-Vega, M.J, Leslie, A.G.W, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a Beta-1- Adrenergic Receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YCZ
 
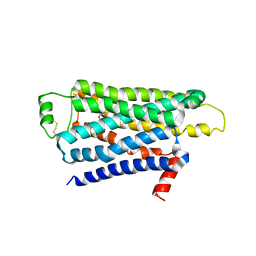 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST IODOCYANOPINDOLOL | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3-iodo-1H-indole-2-carbonitrile, BETA-1 ADRENERGIC RECEPTOR, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Moukhametzianov, R, Warne, T, Edwards, P.C, Serrano-Vega, M.J, Leslie, A.G.W, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a {Beta}1- Adrenergic Receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7O8K
 
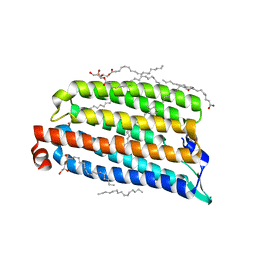 | | NmHR light state structure at 300 us after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8M
 
 | | NmHR light state structure at 2.5 ms (0 - 5 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8Q
 
 | | NmHR light state structure at 22.5 ms (20 - 25 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8Z
 
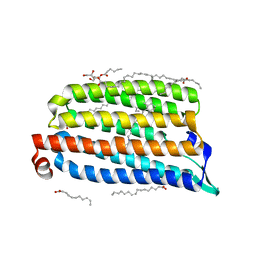 | | Anomalous bromide substructure of NmHR under continuous illumination determined at 13.7 keV with serial crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BROMIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8P
 
 | | NmHR light state structure at 17.5 ms (15 - 20 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8T
 
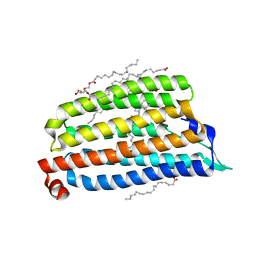 | | NmHR light state structure at 37.5 ms (35 - 40 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8N
 
 | | NmHR light state structure at 7.5 ms (5 - 10 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8S
 
 | | NmHR light state structure at 32.5 ms (30 - 35 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8F
 
 | | NmHR dark state structure determined by serial femtosecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8H
 
 | | NmHR light state structure at 10 ns after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8Y
 
 | | Anomalous bromide substructure of NmHR under dark state conditions determined at 13.7 keV with serial crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BROMIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8I
 
 | | NmHR light state structure at 1 us after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
