7ML2
 
 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7Y3F
 
 | | Structure of the Anabaena PSI-monomer-IsiA supercomplex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Nagao, R, Kato, K, Hamaguchi, T, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2022-06-10 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nat Commun, 14, 2023
|
|
7ML4
 
 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MEI
 
 | | Composite structure of EC+EC | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7EDA
 
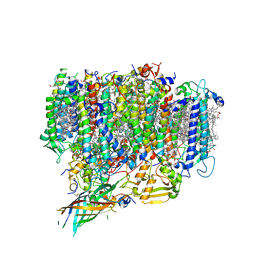 | | Structure of monomeric photosystem II | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Yu, H, Hamaguchi, T, Nakajima, Y, Kato, K, kawakami, K, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Cryo-EM structure of monomeric photosystem II at 2.78 angstrom resolution reveals factors important for the formation of dimer.
Biochim Biophys Acta Bioenerg, 1862, 2021
|
|
7MKA
 
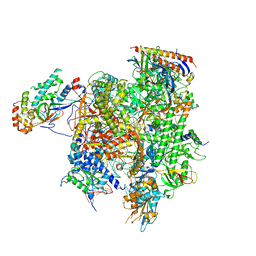 | | Structure of EC+EC (leading EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MK9
 
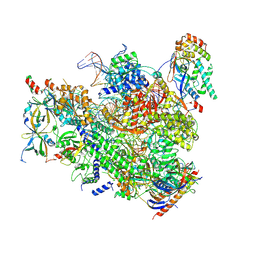 | | Complex structure of trailing EC of EC+EC (trailing EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
3WKL
 
 | |
1WOK
 
 | | Crystal structure of catalytic domain of human poly(ADP-ribose) polymerase complexed with a quinoxaline-type inhibitor | | Descriptor: | 3-(4-CHLOROPHENYL)QUINOXALINE-5-CARBOXAMIDE, Poly [ADP-ribose] polymerase-1 | | Authors: | Iwashita, A, Hattori, K, Yamamoto, H, Ishida, J, Kido, Y, Kamijo, K, Murano, K, Miyake, H, Kinoshita, T, Warizaya, M, Ohkubo, M, Matsuoka, N, Mutoh, S. | | Deposit date: | 2004-08-20 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of quinazolinone and quinoxaline derivatives as potent and selective poly(ADP-ribose) polymerase-1/2 inhibitors.
Febs Lett., 579, 2005
|
|
8GHN
 
 | |
8GHA
 
 | | Hir3 Arm/Tail, Hir2 WD40, C-terminal Hpc2 | | Descriptor: | Histone promoter control protein 2, Histone transcription regulator 3, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
8GHM
 
 | | Hir1 WD40 domains and Asf1/H3/H4 | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
8GHL
 
 | | the Hir complex core | | Descriptor: | Histone transcription regulator 3, Protein HIR1, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
4XNN
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Daphnia pulex | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL | | Authors: | Ebrahim, A, Hobdey, S.E, Podkaminer, K, Taylor II, L.E, Beckham, G.T, Decker, S.R, Himmel, M.E, Cragg, S.M, McGeehan, J.E. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a GH7 Family Cellobiohydrolase from Daphnia pulex
To Be Published
|
|
3VHE
 
 | | Crystal structure of human VEGFR2 kinase domain with a novel pyrrolopyrimidine inhibitor. | | Descriptor: | 1-{2-fluoro-4-[(5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy]phenyl}-3-[3-(trifluoromethyl)phenyl]urea, Vascular endothelial growth factor receptor 2 | | Authors: | Oguro, Y, Miyamoto, N, Okada, K, Takagi, T, Iwata, H, Awazu, Y, Miki, H, Hori, A, Kamiyama, K, Imanura, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, synthesis, and evaluation of 5-methyl-4-phenoxy-5H-pyrrolo[3,2-d]pyrimidine derivatives: novel VEGFR2 kinase inhibitors binding to inactive kinase conformation.
Bioorg.Med.Chem., 18, 2010
|
|
3WFL
 
 | | Crtstal structure of glycoside hydrolase family 5 beta-mannanase from Talaromyces trachyspermus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Suzuki, K, Ichinose, H, Kamino, K, Ogasawara, W, Kaneko, S, Fushinobu, S. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Purification, cloning, functional expression, structure, and characterization of a thermostable beta-mannanase from Talaromyces trachyspermus and its efficiency in production of mannooligosaccharides from coffee wastes
To be Published
|
|
1I6V
 
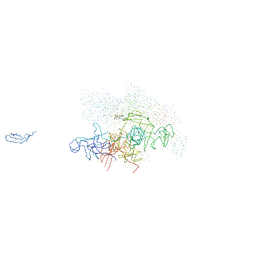 | | THERMUS AQUATICUS CORE RNA POLYMERASE-RIFAMPICIN COMPLEX | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, MAGNESIUM ION, RIFAMPICIN, ... | | Authors: | Campbell, E.A, Korzheva, N, Mustaev, A, Murakami, K, Goldfarb, A, Darst, S.A. | | Deposit date: | 2001-03-05 | | Release date: | 2001-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural mechanism for rifampicin inhibition of bacterial rna polymerase.
Cell(Cambridge,Mass.), 104, 2001
|
|
7KPW
 
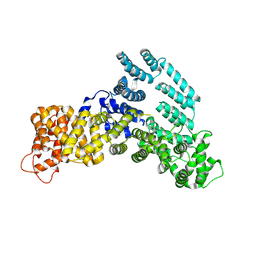 | | Structure of the H-lobe of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12 | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7KPX
 
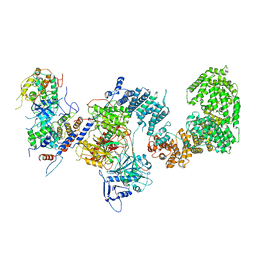 | | Structure of the yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7KPV
 
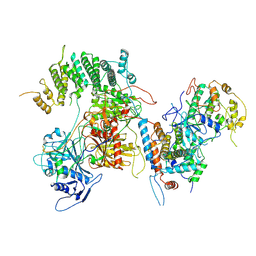 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7JTH
 
 | |
7K01
 
 | | Structure of TFIIH in TFIIH/Rad4-Rad23-Rad33 DNA opening complex | | Descriptor: | DNA repair helicase RAD25, DNA repair helicase RAD3, General transcription and DNA repair factor IIH subunit SSL1, ... | | Authors: | van Eeuwen, T, Min, J.H, Murakami, K. | | Deposit date: | 2020-09-02 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
7K04
 
 | | Structure of TFIIH/Rad4-Rad23-Rad33/DNA in DNA opening | | Descriptor: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | Authors: | van Eeuwen, T, Min, J.H, Murakami, K. | | Deposit date: | 2020-09-03 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
7KUE
 
 | | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin CCL1, ... | | Authors: | van Eeuwen, T, Murakami, K, Li, T, Tsai, K.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
Sci Adv, 7, 2021
|
|
5JGB
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 10 | | Descriptor: | N-(2-methoxy-4-{[3-(4-methylpiperazin-1-yl)propyl]carbamoyl}phenyl)-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
