8D5H
 
 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin
To Be Published
|
|
8EZR
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
6VOP
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli | | Descriptor: | Aldolase | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli
To Be Published
|
|
6VOQ
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae | | Descriptor: | Aldolase, CHLORIDE ION, ZINC ION | | Authors: | Stogios, P.J, Evdokimova, E, McChesney, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae
To Be Published
|
|
6VTV
 
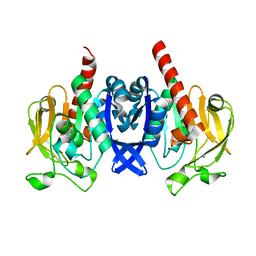 | | Crystal structure of PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase from E. coli | | Descriptor: | Gamma-glutamyl-gamma-aminobutyrate hydrolase PuuD, MANGANESE (II) ION | | Authors: | Stogios, P.J, EVDOKIMOVA, E, DI LEO, R, SAVCHENKO, A, JOACHIMIAK, A, SATCHELL, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-13 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase
To Be Published
|
|
6VU7
 
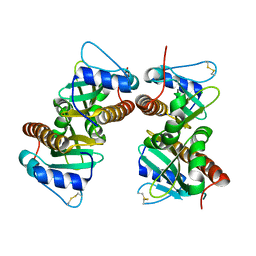 | | Crystal structure of YbjN, a putative transcription regulator from E. coli | | Descriptor: | CHLORIDE ION, YbjN protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of YbjN, a putative transcription regulator from E. coli
To Be Published
|
|
6WOP
 
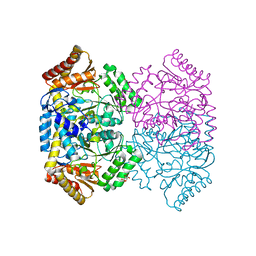 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii | | Descriptor: | 4-aminobutyrate transaminase, CHLORIDE ION, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii
To Be Published
|
|
6WJ8
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Klebsiella pneumoniae in complex with PLP | | Descriptor: | 4-aminobutyrate aminotransferase PuuE | | Authors: | Stogios, P.J, Evdokimova, E, McChesney, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Klebsiella pneumoniae in complex with PLP
To Be Published
|
|
6WQB
 
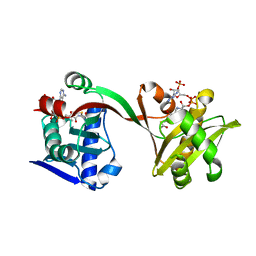 | | Crystal structure of VipF from Legionella hackeliae in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, N-terminal acetyltransferase, GNAT family | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Sandoval, J, Di Leo, R, Savchenko, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of VipF from Legionella hackeliae in complex with acetyl-CoA
To Be Published
|
|
6WQC
 
 | | Crystal structure of VipF from Legionella hackeliae in complex with CoA | | Descriptor: | COENZYME A, N-terminal acetyltransferase, GNAT family | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Sandoval, J, Di Leo, R, Savchenko, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of VipF from Legionella hackeliae in complex with CoA
To Be Published
|
|
4PVA
 
 | | Crystal structure of GH62 hydrolase from thermophilic fungus Scytalidium thermophilum | | Descriptor: | GH62 hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Nocek, B, Kaur, A.P, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2014-03-15 | | Release date: | 2014-11-19 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Functional and structural diversity in GH62 alpha-L-arabinofuranosidases from the thermophilic fungus Scytalidium thermophilum.
Microb Biotechnol, 8, 2015
|
|
7RCA
 
 | | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae | | Descriptor: | CHLORIDE ION, Chorismate synthase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae
To Be Published
|
|
7RGN
 
 | | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris | | Descriptor: | Fructose-bisphosphate aldolase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris
To Be Published
|
|
7RGE
 
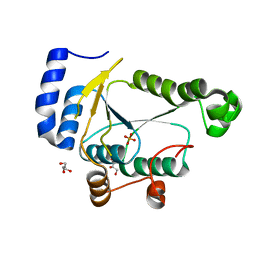 | | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex | | Descriptor: | 3'-phosphoadenylylsulfate reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex
To Be Published
|
|
7REU
 
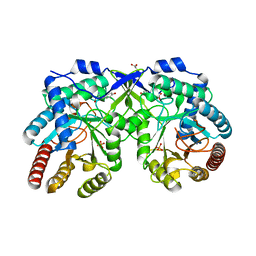 | | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex
To Be Published
|
|
7RH8
 
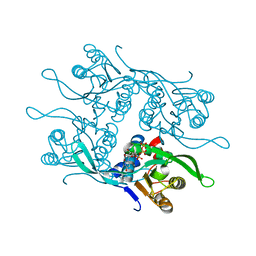 | | Crystal structure of Fur1p from Candida albicans, in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, Uracil phosphoribosyltransferase | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of Fur1p from Candida albicans, in complex with UTP
To Be Published
|
|
7RJ1
 
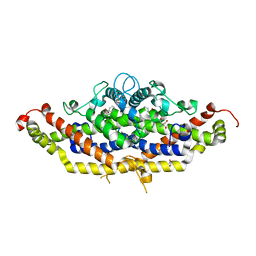 | | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chorismate mutase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp
To Be Published
|
|
7RLL
 
 | | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate | | Descriptor: | Arf3p, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Stogios, P.J, Michalska, K, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-25 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate
To Be Published
|
|
7RQG
 
 | | Crystal structure of the Nsp3 Y3 domain from SARS-CoV-2 | | Descriptor: | Non-structural protein 3 | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the Nsp3 Y3 domain from SARS-CoV-2
To Be Published
|
|
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Sul3 | | Authors: | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2I
 
 | | Crystal structure of sulfonamide resistance enzyme Sul1 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2K
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 in complex with 7,8-dihydropteroate, magnesium, and pyrophosphate | | Descriptor: | 4-AMINOBENZOIC ACID, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
5UB8
 
 | | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Likely rab family GTP-binding protein, ZINC ION | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II)
To Be Published
|
|
5UE7
 
 | | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state
To Be Published
|
|
